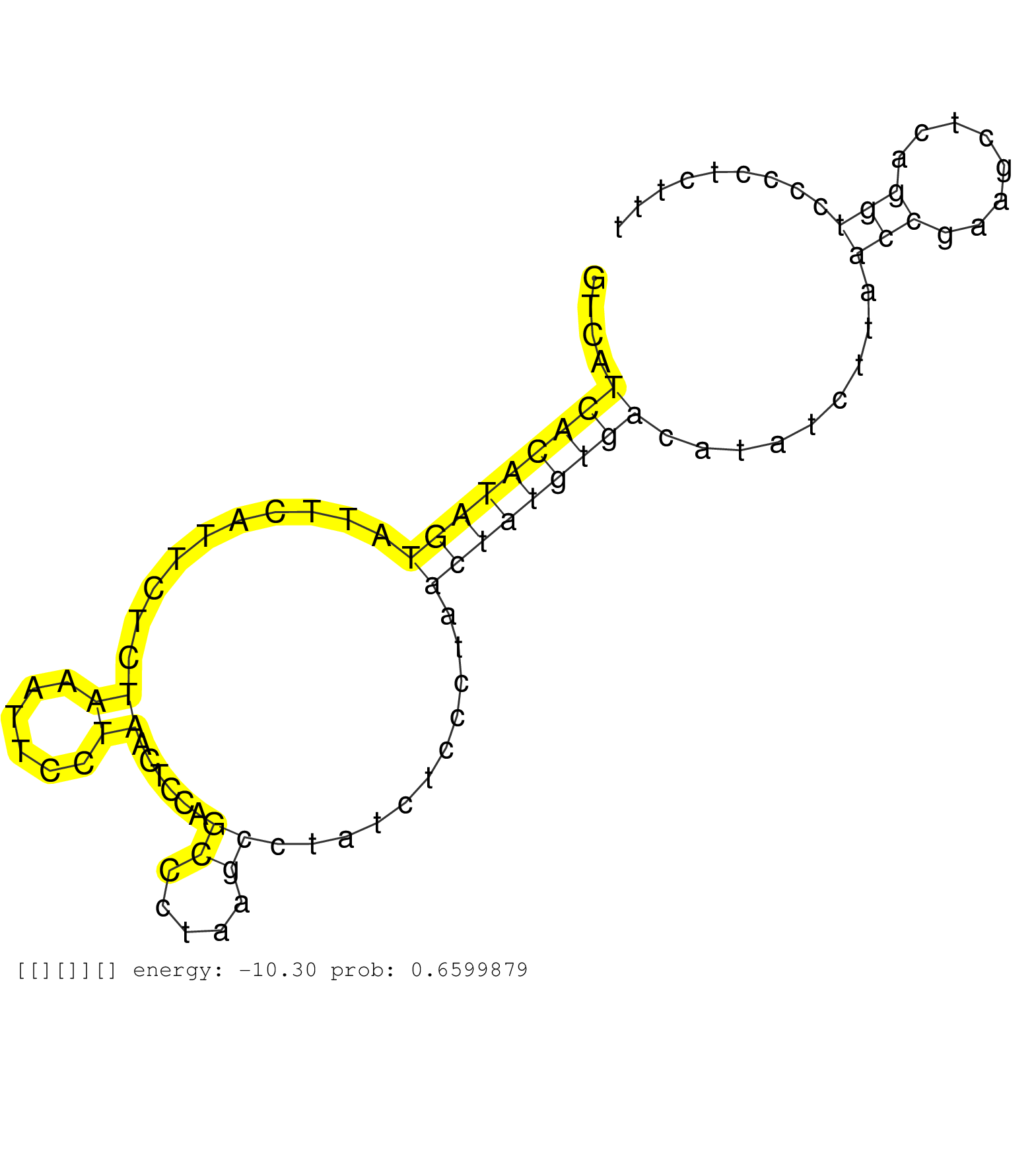

| Gene: 9030025P20Rik | ID: uc008anp.1_intron_2_0_chr17_15117475_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| CAAGTTAAGGATGTCTTTTTCATCTTTGGCCTAATGACACAAGCAAGCAGGTAGATACACGTAGTTACTTTAACAGAAGTTATATTTTATTGTGCCAAGGGTCATCACATAGTATTCATTCTCTAAATTCCTAACTCCAGCCCTAAGCCTATCTCCCTAACTATGTGACATATCTTAACCGAAGCTCAGGTCCCCTCTTTAACAGAAGTCTCTGATTTGAGGGTCAGGATTCTAGGGAGACTTTCCAGCT ........................................................................................................(((((((((..........((......))......((.....))...........))))))))).........(((........)))........................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037900(GSM510436) testes_rep1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGATACACGTAGTTACTTTAACAGAAGTTATATTTTATTGTGCCAAGGGT.................................................................................................................................................... | 52 | 3 | 29.67 | 29.67 | 29.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TCTTTGGCCTAATGACACAAGCAAGCAG........................................................................................................................................................................................................ | 28 | 3 | 1.33 | 1.33 | - | - | - | - | 0.67 | - | 0.33 | - | - | - | 0.33 | - | - | - | - | - |
| ........................TTTGGCCTAATGACACAAGCAAGCAGtt...................................................................................................................................................................................................... | 28 | tt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................AGCAAGCAGGTAGATAttc.............................................................................................................................................................................................. | 19 | ttc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TAATGACACAAGCAAaatg........................................................................................................................................................................................................ | 19 | aatg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TCTTTGGCCTAATGACACAAGCAAGCAGtt...................................................................................................................................................................................................... | 30 | tt | 0.67 | 1.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - |
| ................TTTTCATCTTTGGCCTAATGACACAAGC.............................................................................................................................................................................................................. | 28 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | 0.33 | - | - | - |
| .................TTTCATCTTTGGCCTAATGACACAAGC.............................................................................................................................................................................................................. | 27 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - |
| ..................................TGACACAAGCAAGCAGGTAGATACACGTA........................................................................................................................................................................................... | 29 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| .................TTTCATCTTTGGCCTAATGACACAAGCA............................................................................................................................................................................................................. | 28 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - |
| .....TAAGGATGTCTTTTTCATCTTTGGCCT.......................................................................................................................................................................................................................... | 27 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................AAGCAAGCAGGTAGATACACGTA........................................................................................................................................................................................... | 23 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| ..................TTCATCTTTGGCCTAATGACACAAGCAAGC.......................................................................................................................................................................................................... | 30 | 3 | 0.33 | 0.33 | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - |
| ................TTTTCATCTTTGGCCTAATGACACAAGCA............................................................................................................................................................................................................. | 29 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| CAAGTTAAGGATGTCTTTTTCATCTTTGGCCTAATGACACAAGCAAGCAGGTAGATACACGTAGTTACTTTAACAGAAGTTATATTTTATTGTGCCAAGGGTCATCACATAGTATTCATTCTCTAAATTCCTAACTCCAGCCCTAAGCCTATCTCCCTAACTATGTGACATATCTTAACCGAAGCTCAGGTCCCCTCTTTAACAGAAGTCTCTGATTTGAGGGTCAGGATTCTAGGGAGACTTTCCAGCT ........................................................................................................(((((((((..........((......))......((.....))...........))))))))).........(((........)))........................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037900(GSM510436) testes_rep1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|