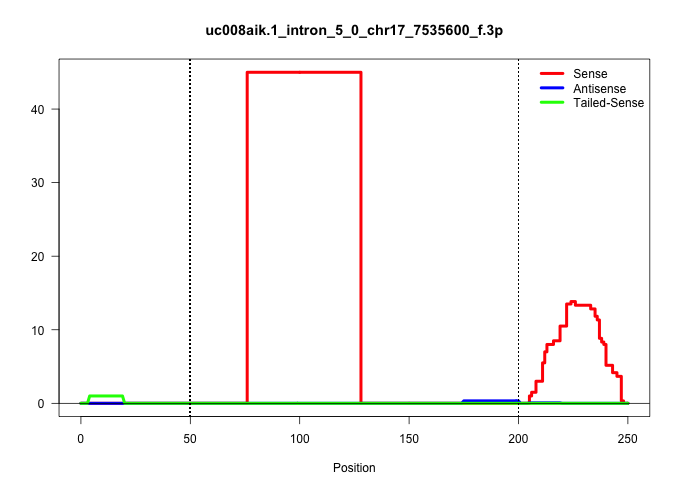
| Gene: Tcp10b | ID: uc008aik.1_intron_5_0_chr17_7535600_f.3p | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(2) PIWI.ip |
(1) TDRD1.ip |
(15) TESTES |
| ACCACATGTAGTGAAGACACAACTAGCAGACAAAGGCTGACTGCACTCAGCACTTCCTGTGTTCCACAGAGCCAGCCACCGGCATTTGCACAGTGACCTCAGGAAACTTAATTCGGACTTGGAACCCTGCACTAGGAATATGACATTTTTTTTCACAGTGTGTGCACAGTCTCTTGAAATCACCTTCTCCACTGTTTCAGACATCTGGTCCTCAAATCCAGAGACTTCGGACGGTGAACTTCTCCTGCAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................CACCGGCATTTGCACAGTGACCTCAGGAAACTTAATTCGGACTTGGAACCCT.......................................................................................................................... | 52 | 2 | 45.00 | 45.00 | 45.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................GACTTCGGACGGTGAACTTCTCCTG... | 25 | 3 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TCAAATCCAGAGACTTCGGACGGTGA............. | 26 | 2 | 1.50 | 1.50 | - | - | - | - | 0.50 | - | 0.50 | 0.50 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGAGACTTCGGACGGTGAACT.......... | 21 | 3 | 1.33 | 1.33 | - | 1.33 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CAAATCCAGAGACTTCGGACGGTGAACT.......... | 28 | 2 | 1.00 | 1.00 | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| ....CATGTAGTGAAGAacc...................................................................................................................................................................................................................................... | 16 | acc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................AAATCCAGAGACTTCGGACGGTGAACTTCT....... | 30 | 2 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CAAATCCAGAGACTTCGGACGGTGA............. | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TCAAATCCAGAGACTTCGGACGGTGAACT.......... | 29 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TCAAATCCAGAGACTTCGGACGGT............... | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ................................................................................................................................................................................................................TCCTCAAATCCAGAGACTTCGGACGGTG.............. | 28 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ........................................................................................................................................................................................................................TCCAGAGACTTCGGACGGTGAACTTCTCC..... | 29 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TCCTCAAATCCAGAGACTTCGGACGGTGA............. | 29 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................GGTCCTCAAATCCAGAGACT........................ | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGGTCCTCAAATCCAGAGACTTCGGACGGT............... | 30 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| .............................................................................................................................................................................................................TGGTCCTCAAATCCAGAGACTTCGGACG................. | 28 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TCCTCAAATCCAGAGACTTCGGACGGTGAA............ | 30 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ...........................................................................................................................................................................................................................AGAGACTTCGGACGGTGAAC........... | 20 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGAGACTTCGGACGGTGAACTTCTCCTG... | 28 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................CTTCGGACGGTGAACTTCTCCTGC.. | 24 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| ACCACATGTAGTGAAGACACAACTAGCAGACAAAGGCTGACTGCACTCAGCACTTCCTGTGTTCCACAGAGCCAGCCACCGGCATTTGCACAGTGACCTCAGGAAACTTAATTCGGACTTGGAACCCTGCACTAGGAATATGACATTTTTTTTCACAGTGTGTGCACAGTCTCTTGAAATCACCTTCTCCACTGTTTCAGACATCTGGTCCTCAAATCCAGAGACTTCGGACGGTGAACTTCTCCTGCAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................GAAATCACCTTCTCCACTGTTTCAGA................................................. | 26 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| .............................................................................................................................................................................................................TGGTCCTCAAATCCA.............................. | 15 | 16 | 0.06 | 0.06 | - | - | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - |