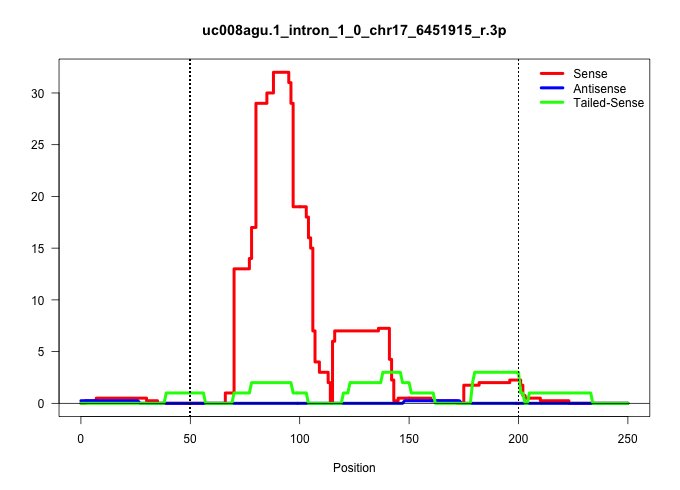
| Gene: Tmem181 | ID: uc008agu.1_intron_1_0_chr17_6451915_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(7) PIWI.ip |
(1) PIWI.mut |
(19) TESTES |
| GATTGATTCCGATCACTGTAAGTCCCATTGGGTGCTTGCTTTCTAGGAACCATCCTTGGCAGGCAGCTGCTGGCGTTCTCTGAGCTTTTGGCGGCGAGTTCTCCCATTCGGCTCATCTGTTGTTGAAATGAGTCGCTCCGGGCTGTGGGTGGCAGGGGTCCTCTTCACTTGCTTTTGTGACGTGGACCTCCTCCTTTCAGAAATGTGCAGAAATCATCGTGGCCCACCTTGGCTACCTGAATTACACACA .................................................................(((((((((.........)))...)))))).......(((...(((((((((.......))..)))))))....)))...((((.((.((((((((...((((........))))...))))).))))).))))................................................... .................................................................66....................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................TGGCGTTCTCTGAGCTTTTGGCGGCGA......................................................................................................................................................... | 27 | 1 | 10.00 | 10.00 | 4.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TGAGCTTTTGGCGGCGAGTTCTCCCA................................................................................................................................................ | 26 | 1 | 8.00 | 8.00 | 4.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TCTGTTGTTGAAATGAGTCGCTCCGG............................................................................................................. | 26 | 1 | 3.00 | 3.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TGGCGTTCTCTGAGCTTTTGGCGGCG.......................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TCTGTTGTTGAAATGAGTCGCTCCGGG............................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TCTGAGCTTTTGGCGGCGAGTTCTCC.................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TGAGCTTTTGGCGGCGAGTTCTCCCAT............................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TGGCGGCGAGTTCTCCCATTCGGCTC........................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................ACGTGGACCTCCTCCTTTCAG.................................................. | 21 | 4 | 1.50 | 1.50 | - | - | - | - | - | - | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TCTGAGCTTTTGGCGGCGAGTTCTCCCAT............................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CTGTTGTTGAAATGAGTCGCTCCGGGC........................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TGAGCTTTTGGCGGCGAGTTCTCCC................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CGGGCTGTGGGTGGCAGGGGTaat........................................................................................ | 24 | aat | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TGGCGTTCTCTGAGCTTTTGGCGGCGt......................................................................................................................................................... | 27 | t | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGTTGAAATGAGTCGCTCCGGGCTGTa....................................................................................................... | 27 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TTTCTAGGAACCATgtag................................................................................................................................................................................................. | 18 | gtag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TGAAATGAGTCGCTCCGGGCTGTGGGTt................................................................................................... | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TTTTGGCGGCGAGTTCTCCCATTCGGCT......................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TCTGTTGTTGAAATGAGTCGCTCCGGGC........................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TCTGAGCTTTTGGCGGCGAGTTCTtc.................................................................................................................................................. | 26 | tc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................CTGCTGGCGTTCTCTGAGCTTTTGGCGGC........................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................CTCTGAGCTTTTGGCGGCGAGTTCTC................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGTGACGTGGACCTCCTCCTTTCAGAA................................................ | 27 | 4 | 1.00 | 1.00 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CGTGGACCTCCTCCTTTCAGAga............................................... | 23 | ga | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TGAGCTTTTGGCGGCGAGTTCTCCCATTC............................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGCAGAAATCATCGTGGCCCACCTTGGta................ | 29 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................ACGTGGACCTCCTCCTTTCAGt................................................. | 22 | t | 0.50 | 1.50 | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | 0.25 | - | - | - | - |
| ...............................................................................................................................................................................TGTGACGTGGACCTCCTCCTTTCAGA................................................. | 26 | 4 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TCCGGGCTGTGGGTGGCAGGGGTC.......................................................................................... | 24 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| .................................................................................................................................................TGGGTGGCAGGGGTCCTCTTCACTTGC.............................................................................. | 27 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - |
| ......................................................................................................................................................................................TGGACCTCCTCCTTTCAGAAATGTGCAG........................................ | 28 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TCAGAAATGTGCAGAAATCATCGTGGC........................... | 27 | 4 | 0.25 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TCCGATCACTGTAAGTCCCATTGGGTGC....................................................................................................................................................................................................................... | 28 | 4 | 0.25 | 0.25 | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGTGACGTGGACCTCCTCCTTTCAGAAA............................................... | 28 | 4 | 0.25 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CGTGGACCTCCTCCTTTCAGAAATGTGCA......................................... | 29 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - |
| ..TTGATTCCGATCACTGTAAGTCCCATTG............................................................................................................................................................................................................................ | 28 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - |
| GATTGATTCCGATCACTGTAAGTCCCATTGGGTGCTTGCTTTCTAGGAACCATCCTTGGCAGGCAGCTGCTGGCGTTCTCTGAGCTTTTGGCGGCGAGTTCTCCCATTCGGCTCATCTGTTGTTGAAATGAGTCGCTCCGGGCTGTGGGTGGCAGGGGTCCTCTTCACTTGCTTTTGTGACGTGGACCTCCTCCTTTCAGAAATGTGCAGAAATCATCGTGGCCCACCTTGGCTACCTGAATTACACACA .................................................................(((((((((.........)))...)))))).......(((...(((((((((.......))..)))))))....)))...((((.((.((((((((...((((........))))...))))).))))).))))................................................... .................................................................66....................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................GGCGAGTTCTCCCATTCGGCTCATCTGT.................................................................................................................................. | 28 | 1 | 5.00 | 5.00 | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGGCAGGCAGCTGCTGGCGTTCTCTGAt....................................................................................................................................................................... | 28 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGGCAGGGGTCCTCTTCACTTGCTTTTGtt......................................................................... | 30 | tt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GTGGCAGGGGTCCTCTTCACTTGCTT............................................................................ | 26 | 4 | 0.25 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |