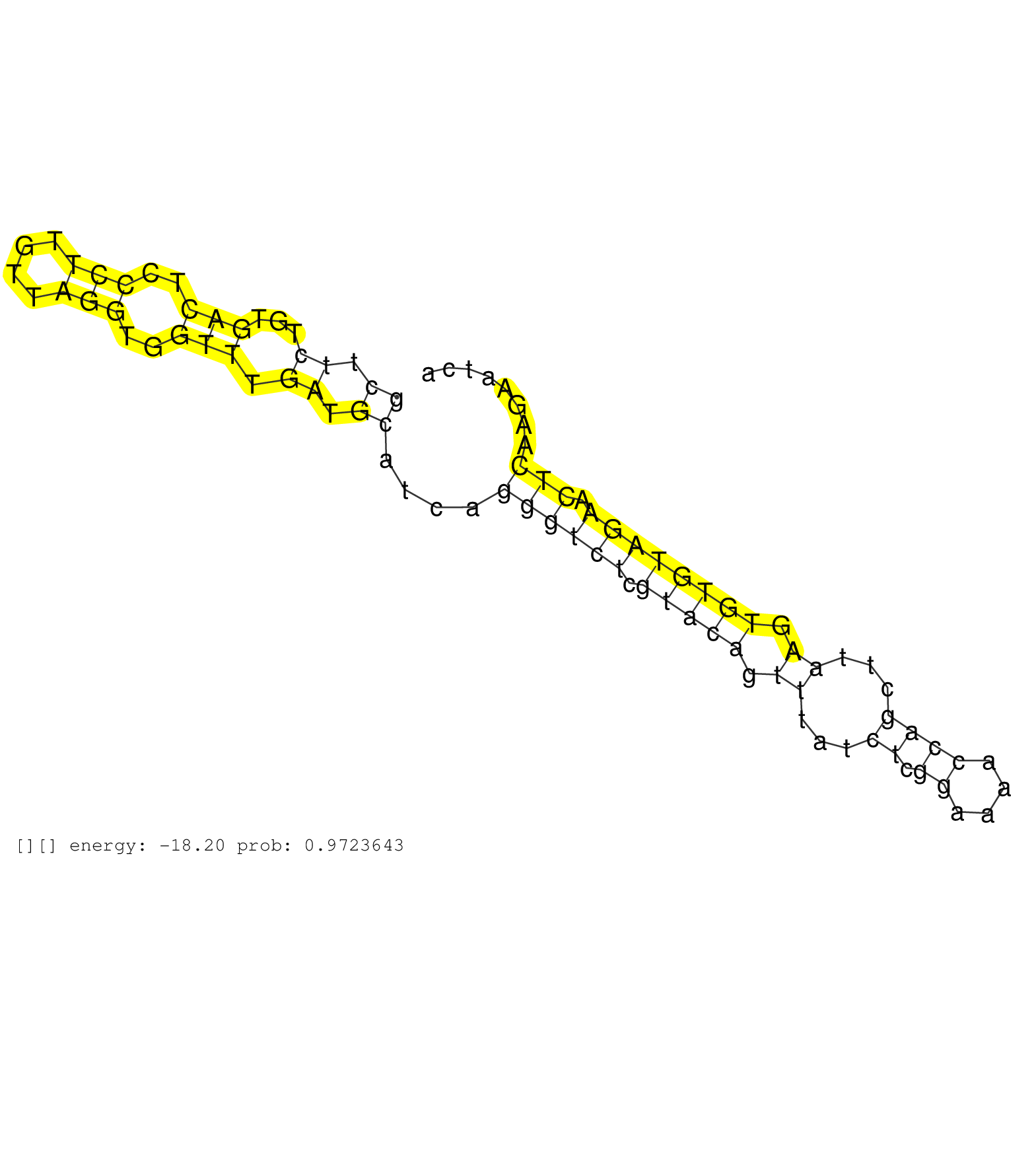
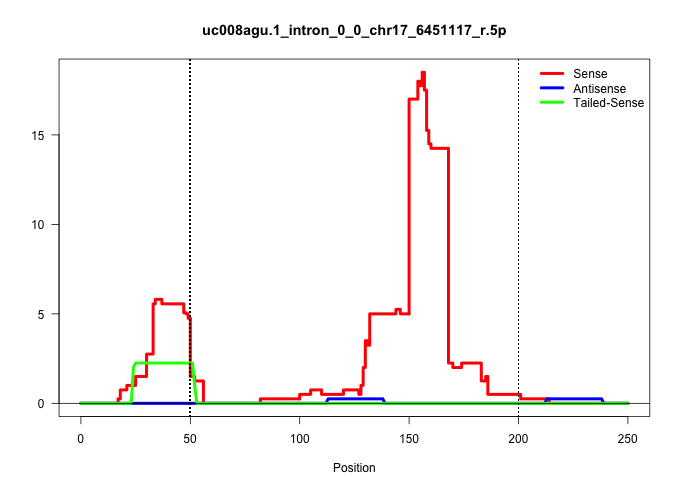
| Gene: Tmem181 | ID: uc008agu.1_intron_0_0_chr17_6451117_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(2) PIWI.mut |
(21) TESTES |
| CTGTGGGGTTTGAGCACCTGAACCAGCCCATCAAGGATATGAACTTCACGGTGAGTGTGCCGTGCAGCCAGCGGCCAGCTTCTGTGACTCCCTTGTTAGGTGGTTTGATGCATCAGGGTCTCGTACAGTTTATCTCGGAAAACCAGCTTAAGTGTGTAGAACTCAAGAATCACTTAATCAAACAGCCCGGACACTGGAGAAGCCAGGGTGCGCTCAGCCAGCCAGCCATCCCACGCTTAGCTAGCTGCCT .............................................................................((.((...(((..(((....)))..))).)).))....((((((.(((((.((...((.((....))))...)).)))))))).)))...................................................................................... .............................................................................78............................................................................................172............................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................AGTGTGTAGAACTCAAGAATCACTTAATCAAACAGCCCGGACACTGGAGAAG................................................ | 52 | 1 | 48.00 | 48.00 | 48.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TCTCGTACAGTTTATCTCGGAAAACCAGCTTAAGTGTGTAGAACTCAAGAAT................................................................................ | 52 | 4 | 11.75 | 11.75 | 11.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................AGGATATGAACTTCACG........................................................................................................................................................................................................ | 17 | 4 | 2.75 | 2.75 | - | - | 1.25 | 0.25 | - | - | - | - | 0.75 | 0.25 | - | - | - | - | - | - | - | - | - | 0.25 | - | - |
| ..................................................................................................................................TATCTCGGAAAACCAGCTTAAGTGTGTA............................................................................................ | 28 | 4 | 1.25 | 1.25 | - | 1.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AAAACCAGCTTAAGTGTGTAGAACTCAAG................................................................................... | 29 | 4 | 1.25 | 1.25 | - | - | - | - | - | 1.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TCAAGGATATGAACTTCACGGTGAGT.................................................................................................................................................................................................. | 26 | 4 | 1.25 | 1.25 | - | 0.25 | - | - | 0.25 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | 0.25 | - | - | - | - |
| ..........................................................................................................................................................TGTAGAACTCAAGAATCACTTAATCAAAC................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......GTTTGAGCACCTGAAaaaa................................................................................................................................................................................................................................ | 19 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................AGCCCATCAAGGATATGAACTTCACGtt...................................................................................................................................................................................................... | 28 | tt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................AGCCCATCAAGGATATGAACTTCACGtgg..................................................................................................................................................................................................... | 29 | tgg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TAGAACTCAAGAATCACTTAATCAAACAGC................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCTCGGAAAACCAGCTTAAGTGTGTA............................................................................................ | 26 | 4 | 0.75 | 0.75 | - | 0.25 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TTATCTCGGAAAACCAGCTTAAGTGTGT............................................................................................. | 28 | 4 | 0.75 | 0.75 | - | 0.25 | - | - | 0.25 | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCTCGGAAAACCAGCTTAAGTGTGTAG........................................................................................... | 27 | 4 | 0.75 | 0.75 | - | 0.25 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................CCCGGACACTGGAGAAGCCA............................................. | 20 | 4 | 0.75 | 0.75 | - | - | - | - | - | - | - | - | - | - | 0.75 | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGAACCAGCCCATCAAGGATATGAACTTC........................................................................................................................................................................................................... | 29 | 4 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................CTGAACCAGCCCATCAAGGA..................................................................................................................................................................................................................... | 20 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - |
| .....................ACCAGCCCATCAAGGATATGAACTTCAC......................................................................................................................................................................................................... | 28 | 4 | 0.25 | 0.25 | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................GCCCATCAAGGATATGAACTTCACGat...................................................................................................................................................................................................... | 27 | at | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| .................................AGGATATGAACTTCACGtgga.................................................................................................................................................................................................... | 21 | tgga | 0.25 | 2.75 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - |
| ..................................................................................TGTGACTCCCTTGTTAGGTGGTTTGATG............................................................................................................................................ | 28 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - |
| .........................GCCCATCAAGGATATGAACTTCACGGT...................................................................................................................................................................................................... | 27 | 4 | 0.25 | 0.25 | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TTTATCTCGGAAAACCAGCTTAAGTGTG.............................................................................................. | 28 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TTTATCTCGGAAAACCAGCTTAAGTGT............................................................................................... | 27 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................CCCGGACACTGGAGAAGCCAGGGTGCGCT.................................... | 29 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |
| .........................................................................................................TGATGCATCAGGGTCTCGTACAGTTT....................................................................................................................... | 26 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TTATCTCGGAAAACCAGCTTAAGTGTGTA............................................................................................ | 29 | 4 | 0.25 | 0.25 | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TGGTTTGATGCATCAGGGTCTCGTACA........................................................................................................................... | 27 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TCTCGGAAAACCAGCTTAAGTGTGTAGA.......................................................................................... | 28 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - |
| ..................................GGATATGAACTTCACG........................................................................................................................................................................................................ | 16 | 4 | 0.25 | 0.25 | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TATCTCGGAAAACCAGCTTAAGTGTGT............................................................................................. | 27 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TAATCAAACAGCCCGGACACTGGAGAA................................................. | 27 | 4 | 0.25 | 0.25 | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGAACCAGCCCATCAAGGATATGAACTTCttt........................................................................................................................................................................................................ | 32 | ttt | 0.25 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - |
| ........................................................................................................................TCGTACAGTTTATCTCGGAAAACCAG........................................................................................................ | 26 | 4 | 0.25 | 0.25 | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................GCCCATCAAGGATATGAACTTCACG........................................................................................................................................................................................................ | 25 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AGCTTAAGTGTGTAGAACTCAAGAAT................................................................................ | 26 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................AGGATATGAACTTCA.......................................................................................................................................................................................................... | 15 | 17 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTGTGGGGTTTGAGCACCTGAACCAGCCCATCAAGGATATGAACTTCACGGTGAGTGTGCCGTGCAGCCAGCGGCCAGCTTCTGTGACTCCCTTGTTAGGTGGTTTGATGCATCAGGGTCTCGTACAGTTTATCTCGGAAAACCAGCTTAAGTGTGTAGAACTCAAGAATCACTTAATCAAACAGCCCGGACACTGGAGAAGCCAGGGTGCGCTCAGCCAGCCAGCCATCCCACGCTTAGCTAGCTGCCT .............................................................................((.((...(((..(((....)))..))).)).))....((((((.(((((.((...((.((....))))...)).)))))))).)))...................................................................................... .............................................................................78............................................................................................172............................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................CAGGGTCTCGTACAGTTTATCTCGGA............................................................................................................... | 26 | 4 | 0.25 | 0.25 | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TCAGCCAGCCAGCCATCCCACGCTTA........... | 26 | 4 | 0.25 | 0.25 | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |