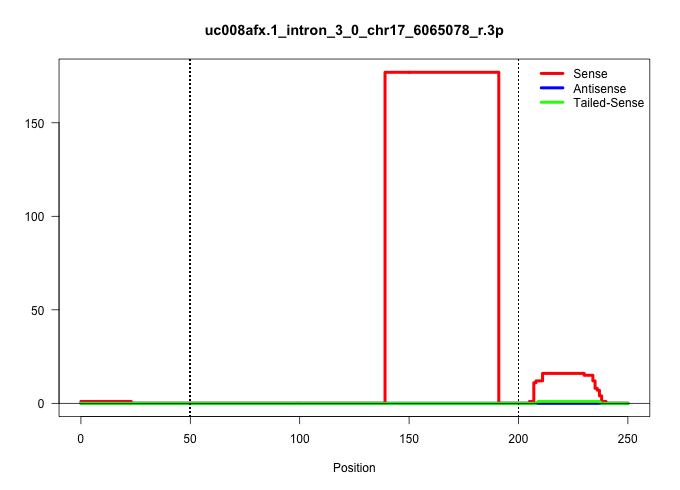
| Gene: Serac1 | ID: uc008afx.1_intron_3_0_chr17_6065078_r.3p | SPECIES: mm9 |
 |
|
(2) PIWI.ip |
(2) PIWI.mut |
(12) TESTES |
| TAGCAGGGGTTGAAGCGTAGAACTAAAAATAAATGGTTATGTAACCTTCACTTTGGTGGGCGAGTATGCTCCACATCACAACCACTGGCCATCCTCTCTCCCTCCTTGCCTGTGTGTATATCCCGGCCCCCGCTCTTTGGAGGATCTTATACTCGGTTGTGGAGTTTGTGTCTAAAGACTTCCTCTGTGTTTTATTCCAGGACTCCTCCACTGAAGAGGAACTCAGGCATCTGCTGGCCTCTCTGCCTCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................GAGGATCTTATACTCGGTTGTGGAGTTTGTGTCTAAAGACTTCCTCTGTGTT........................................................... | 52 | 1 | 177.00 | 177.00 | 177.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CCACTGAAGAGGAACTCAGGCATCTGCT............... | 28 | 1 | 4.00 | 4.00 | - | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................................................CCACTGAAGAGGAACTCAGGCATCTGC................ | 27 | 1 | 3.00 | 3.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................................................................................TGAAGAGGAACTCAGGCATCTGCTGG............. | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................................................................................TGAAGAGGAACTCAGGCATCTGCTGGC............ | 27 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CCACTGAAGAGGAACTCAGGCATCTGCTG.............. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CTCCACTGAAGAGGAACTCAGGCAT.................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CCACTGAAGAGGAACTCAGGCATCTGCTGGC............ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................................................................................................................................CACTGAAGAGGAACTCAGGCATCTGCTGGCCT.......... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................ACTGAAGAGGAACTCAGGCATCTGCTGGt............ | 29 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................................................................................CCACTGAAGAGGAACTCAGGCATCTGCTGG............. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| TAGCAGGGGTTGAAGCGTAGAACTAAAAATAAATGGTTATGTAACCTTCACTTTGGTGGGCGAGTATGCTCCACATCACAACCACTGGCCATCCTCTCTCCCTCCTTGCCTGTGTGTATATCCCGGCCCCCGCTCTTTGGAGGATCTTATACTCGGTTGTGGAGTTTGTGTCTAAAGACTTCCTCTGTGTTTTATTCCAGGACTCCTCCACTGAAGAGGAACTCAGGCATCTGCTGGCCTCTCTGCCTCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|