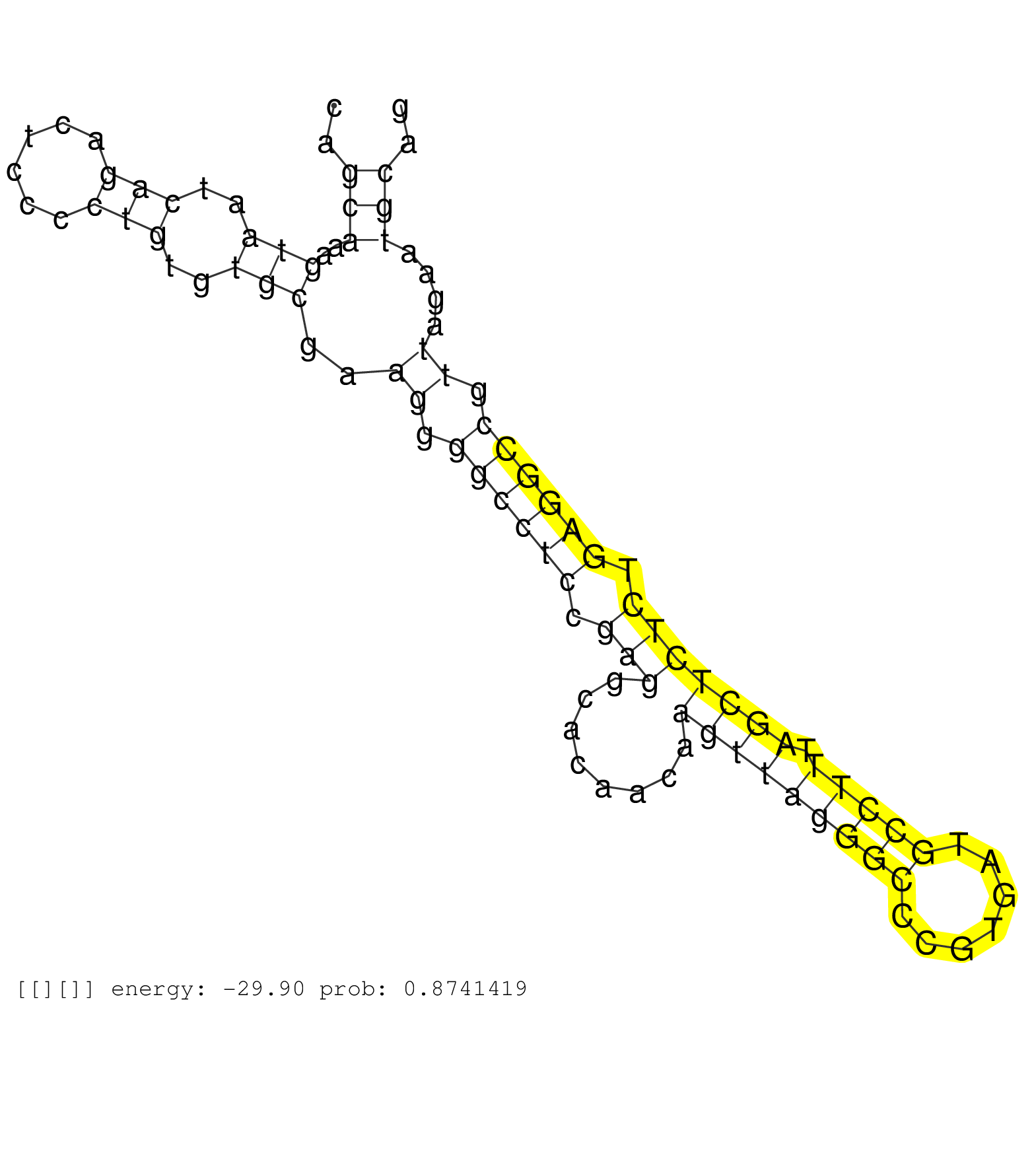
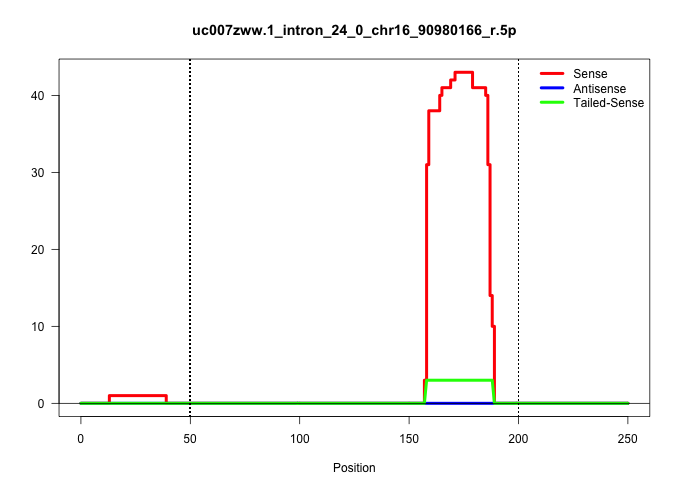
| Gene: Synj1 | ID: uc007zww.1_intron_24_0_chr16_90980166_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
| ATCTGCTTGGGTCTAAGGAAGGCGAGCATATGCTGAGCAAGGCTTTCCAGGTGAGTGTGACCACTGTCGTCTGTCTCTTCTGCTCACCCAGAGTGGAACCCAGCAAAGTAATCAGACTCCCCTGTGTGCGAAGGGGCCTCCGAGGCACAACAAGTTAGGGCCCGTGATGCCTTTAGCTCTCTGAGGCCGTTAGAATGCAGTATAGGCAAAATAATTAATCAGTTTTGGATCGATGTAAATTTTGTTATGT ......................................................................................................(((..(((..(((......)))..)))..((.((((((.(((........(((((((((.......))))).))))))).)))))).))....))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475281(GSM475281) total RNA. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesWT3() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................GGCCCGTGATGCCTTTAGCTCTCTGAGGC............................................................... | 29 | 1 | 22.00 | 22.00 | 3.00 | 9.00 | 1.00 | - | 2.00 | - | - | - | 3.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - |
| .............................................................................................................................................................GGGCCCGTGATGCCTTTAGCTC....................................................................... | 22 | 1 | 14.00 | 14.00 | 13.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GGCCCGTGATGCCTTTAGCTCTCTGAGG................................................................ | 28 | 1 | 8.00 | 8.00 | - | 1.00 | 1.00 | 2.00 | - | - | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................GGCCCGTGATGCCTTTAGCTC....................................................................... | 21 | 1 | 6.00 | 6.00 | 5.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GGCCCGTGATGCCTTTAGCTCTCTGAGGCC.............................................................. | 30 | 1 | 6.00 | 6.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GCCCGTGATGCCTTTAGCTCTCTGAGGCCG............................................................. | 30 | 1 | 5.00 | 5.00 | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 |
| ...............................................................................................................................................................GCCCGTGATGCCTTTAGCTCTCTGAGGCC.............................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................GGGCCCGTGATGCCTTTAGCTCTCTGAGGCCG............................................................. | 32 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................................................................................................GATGCCTTTAGCTCTCTGAGGCCG............................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................GGGCCCGTGATGCCTTTAGCTCTCTGAG................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TTTAGCTCTCTGAGGCCG............................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GGCCCGTGATGCCTTTAGCTCTCTGAGGCCG............................................................. | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TAAGGAAGGCGAGCATATGCTGAGCA................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................GGCCCGTGATGCCTTTAGCTCTCTGAGGCag............................................................. | 31 | ag | 1.00 | 22.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................CCTTTAGCTCTCTGAGGCCG............................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GGCCCGTGATGCCTTTAGCTCTCTGAGGacg............................................................. | 31 | acg | 1.00 | 8.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GCCCGTGATGCCTTTAGCTCTCTGAGGC............................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GGCCCGTGATGCCTTTAGCTCTCTGAGGCCc............................................................. | 31 | c | 1.00 | 6.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGATGCCTTTAGCTCTCTGAGG................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGATGCCTTTAGCTCTCTGAGGCC.............................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ATCTGCTTGGGTCTAAGGAAGGCGAGCATATGCTGAGCAAGGCTTTCCAGGTGAGTGTGACCACTGTCGTCTGTCTCTTCTGCTCACCCAGAGTGGAACCCAGCAAAGTAATCAGACTCCCCTGTGTGCGAAGGGGCCTCCGAGGCACAACAAGTTAGGGCCCGTGATGCCTTTAGCTCTCTGAGGCCGTTAGAATGCAGTATAGGCAAAATAATTAATCAGTTTTGGATCGATGTAAATTTTGTTATGT ......................................................................................................(((..(((..(((......)))..)))..((.((((((.(((........(((((((((.......))))).))))))).)))))).))....))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475281(GSM475281) total RNA. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesWT3() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|