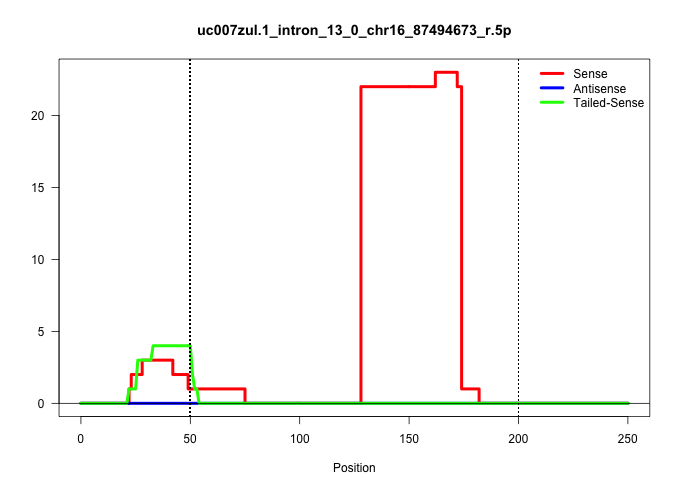
| Gene: Cct8 | ID: uc007zul.1_intron_13_0_chr16_87494673_r.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(10) TESTES |
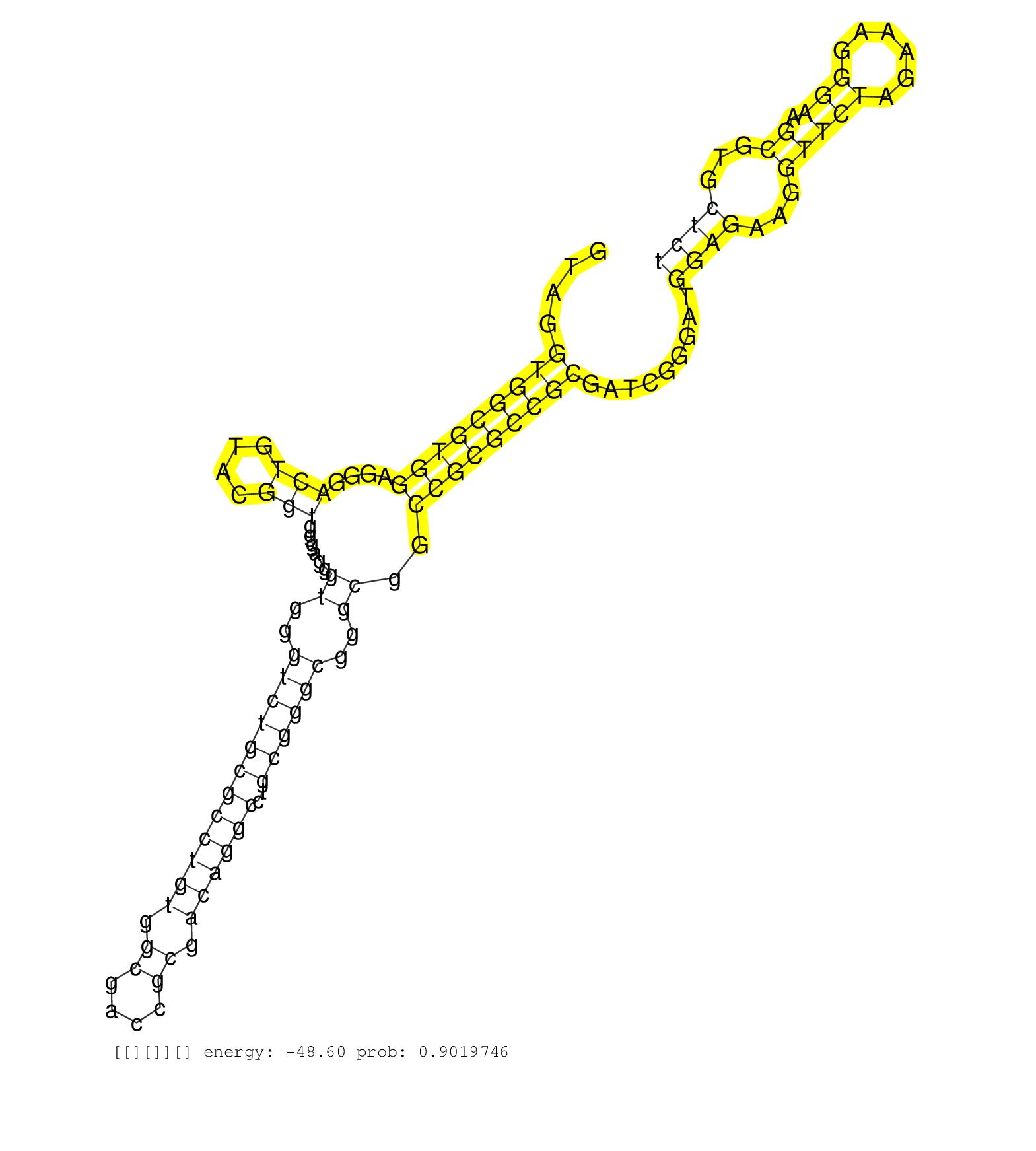
| ACGTCCCCAAGGCCCCGGGCTTTGCCCAGATGCTCAAGGATGGAGCCAAAGTAGGTGGCGTGGAGGGACTGTACGGTGGGAGGGTGGGTCTGCGCCTGTGGCGACCGCGACAGGCCTGCGGGCGGGCGGCCGCGCCGCGATCGGGATGGAGAAGGTTCTAGAAAGGGAAGCGTGCTCTGGATGCCGGCTCGCTGGAACTGGTGGGCATTTCTGTTATTCCCCCGGCGGCTTCGGGTTTGCATCCCAGCTC ......................................................(((((((((....(((....)))......((..((((((((((((.((....)).))))))..))))))..))..))))))))).........((((...(((((......))).))...))))........................................................................ ..................................................51.............................................................................................................................178...................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................GCCGCGCCGCGATCGGGATGGAGAAGGTTCTAGAAAGGGAAGCGTGCTCTGG...................................................................... | 52 | 1 | 65.00 | 65.00 | 65.00 | - | - | - | - | - | - | - | - | - |
| .................................................AGTAGGTGGCGTGGAGGGACTGTACGGT............................................................................................................................................................................. | 28 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................GCCTGCGGGCGGGCGaa........................................................................................................................ | 17 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................GGGCGGCCGCGCCGCGATCGGGATGGAGAAGGT.............................................................................................. | 33 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................TCAAGGATGGAGCCAAAca...................................................................................................................................................................................................... | 19 | ca | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................GCCCAGATGCTCAAGGATG................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................CAGATGCTCAAGGATGGAGCCAAAc....................................................................................................................................................................................................... | 25 | c | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................GCCAAAGTAGGTGGCGTGGAGGGACTGTACGGTGGG.......................................................................................................................................................................... | 36 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................CTTCGGGTTTGCATCCCAGC.. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................TGCCCAGATGCTCAAGGATGGAGCCAAAc....................................................................................................................................................................................................... | 29 | c | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................AAGTAGGTGGCGTGGAGGGACTGTACG............................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................................AAGGGAAGCGTGCTCTGGAT.................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................CAGATGCTCAAGGATGGAGCCAAAcatt.................................................................................................................................................................................................... | 28 | catt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................GATGCTCAAGGATGGAGCCA.......................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................AGGATGGAGCCAAAGTAGG................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................TGCCCAGATGCTCAAGGATGGAGCCAA......................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ACGTCCCCAAGGCCCCGGGCTTTGCCCAGATGCTCAAGGATGGAGCCAAAGTAGGTGGCGTGGAGGGACTGTACGGTGGGAGGGTGGGTCTGCGCCTGTGGCGACCGCGACAGGCCTGCGGGCGGGCGGCCGCGCCGCGATCGGGATGGAGAAGGTTCTAGAAAGGGAAGCGTGCTCTGGATGCCGGCTCGCTGGAACTGGTGGGCATTTCTGTTATTCCCCCGGCGGCTTCGGGTTTGCATCCCAGCTC ......................................................(((((((((....(((....)))......((..((((((((((((.((....)).))))))..))))))..))..))))))))).........((((...(((((......))).))...))))........................................................................ ..................................................51.............................................................................................................................178...................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|