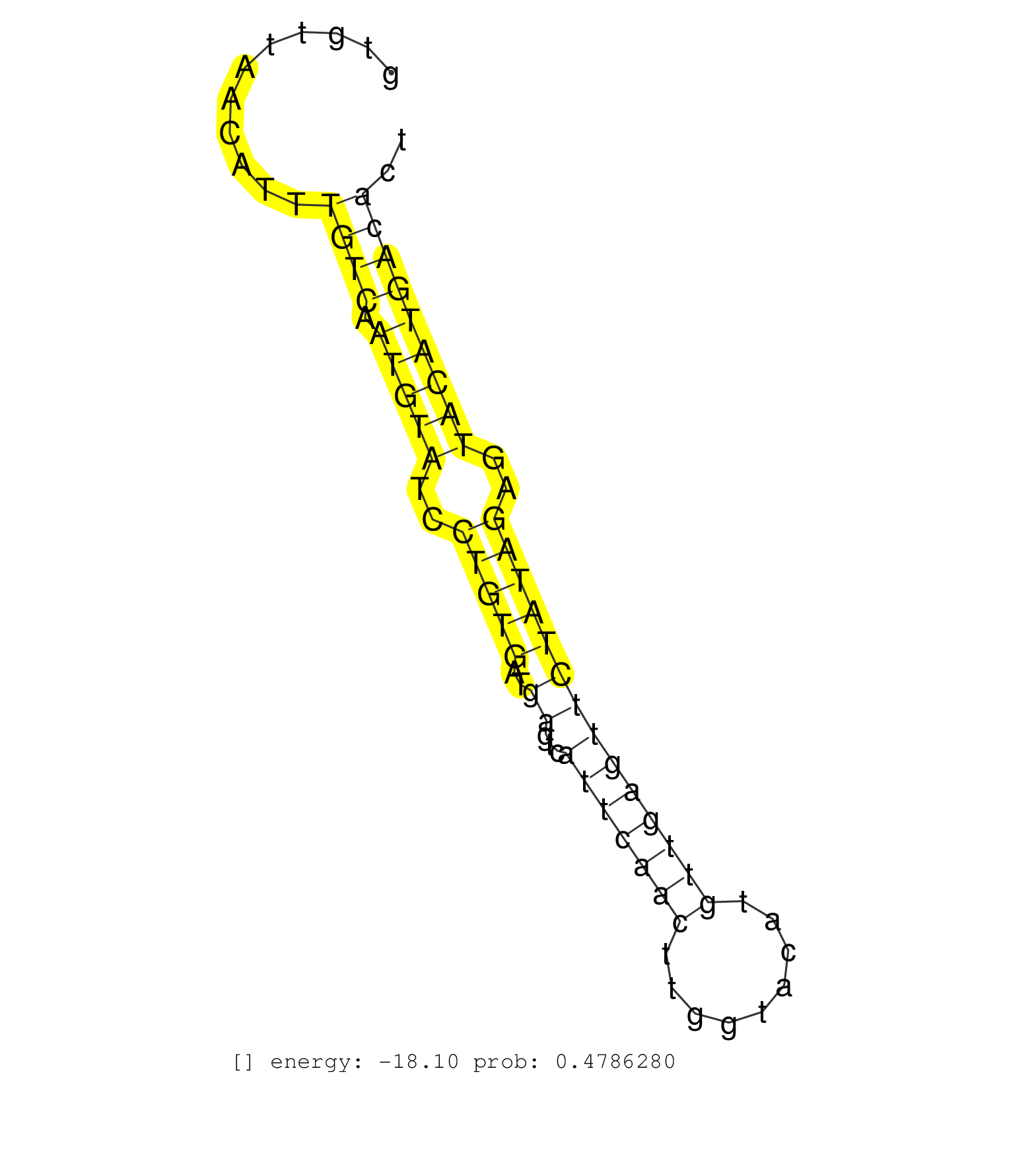
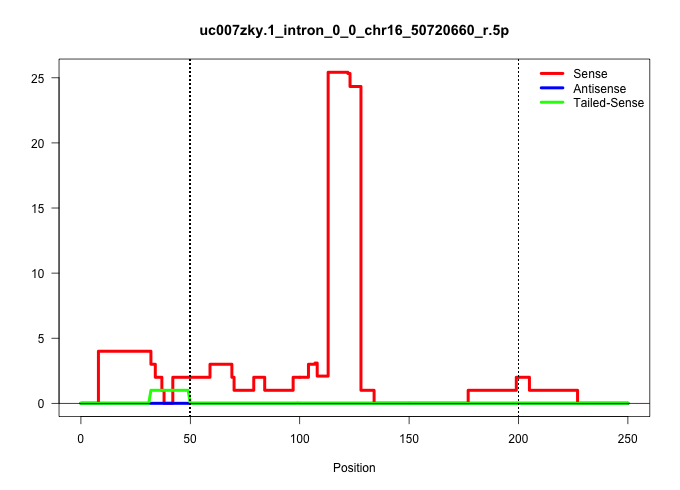
| Gene: AK151508 | ID: uc007zky.1_intron_0_0_chr16_50720660_r.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(15) TESTES |
| GGTAGATATACAAGAAGAACTGCATTGGGATAACCGCCATCTTACAAAGGGTCAGTGTTAACATTTGTCAATGTATCCTGTGATGAGTCATTCAACTTGGTACATGTTGAGTTCTATAGAGTACATGACACTCTTATGCCTGTGAGCAAGCCATAATGTAAAATAATTCATCCAAACCCATAGGAGTGTGTGTCTTTTGTAAGGCACGCATCAGAATATTTACACACTCATGTCATAAACTGGATCACTG .................................................................((((.(((((..(((((..((...(((((((.........))))))))))))))..)))))))))........................................................................................................................ ......................................................55...........................................................................132.................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................CTATAGAGTACATGACACTCTTATGCCTGTGAGCAAGCCATAATGTAAAATA..................................................................................... | 52 | 1 | 137.00 | 137.00 | 137.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TTATGCCTGTGAGCAAGCCATAA.............................................................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AACATTTGTCAATGTATCCTGTGAT...................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................TGGTACATGTTGAGTTCTATAGAGTA............................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........TACAAGAAGAACTGCATTGGGATAAC........................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................GTGATGAGTCATTCAACTTGGTACATGTT.............................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................GGTACATGTTGAGTTCTATAGAGT................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........TACAAGAAGAACTGCATTGGGATA.......................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................TACAAAGGGTCAGTGTTAACATTTGTC..................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........TACAAGAAGAACTGCATTGGGATAACCGC..................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TGTTGAGTTCTATAGAGTACATGACACTCT.................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................TACAAAGGGTCAGTGTTAACATTTGTCA.................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........TACAAGAAGAACTGCATTGGGATAACCGCC.................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................ACCGCCATCTTACAgagc........................................................................................................................................................................................................ | 18 | gagc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................ACTCATGTCATAAACTGGAT..... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................TAAGGCACGCATCAGAATATTTACACAC....................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CCATAGGAGTGTGTGTCTTTTGTAAGGC............................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................TGAGTTCTATAGAGT................................................................................................................................ | 15 | 11 | 0.09 | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.09 |
| GGTAGATATACAAGAAGAACTGCATTGGGATAACCGCCATCTTACAAAGGGTCAGTGTTAACATTTGTCAATGTATCCTGTGATGAGTCATTCAACTTGGTACATGTTGAGTTCTATAGAGTACATGACACTCTTATGCCTGTGAGCAAGCCATAATGTAAAATAATTCATCCAAACCCATAGGAGTGTGTGTCTTTTGTAAGGCACGCATCAGAATATTTACACACTCATGTCATAAACTGGATCACTG .................................................................((((.(((((..(((((..((...(((((((.........))))))))))))))..)))))))))........................................................................................................................ ......................................................55...........................................................................132.................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|