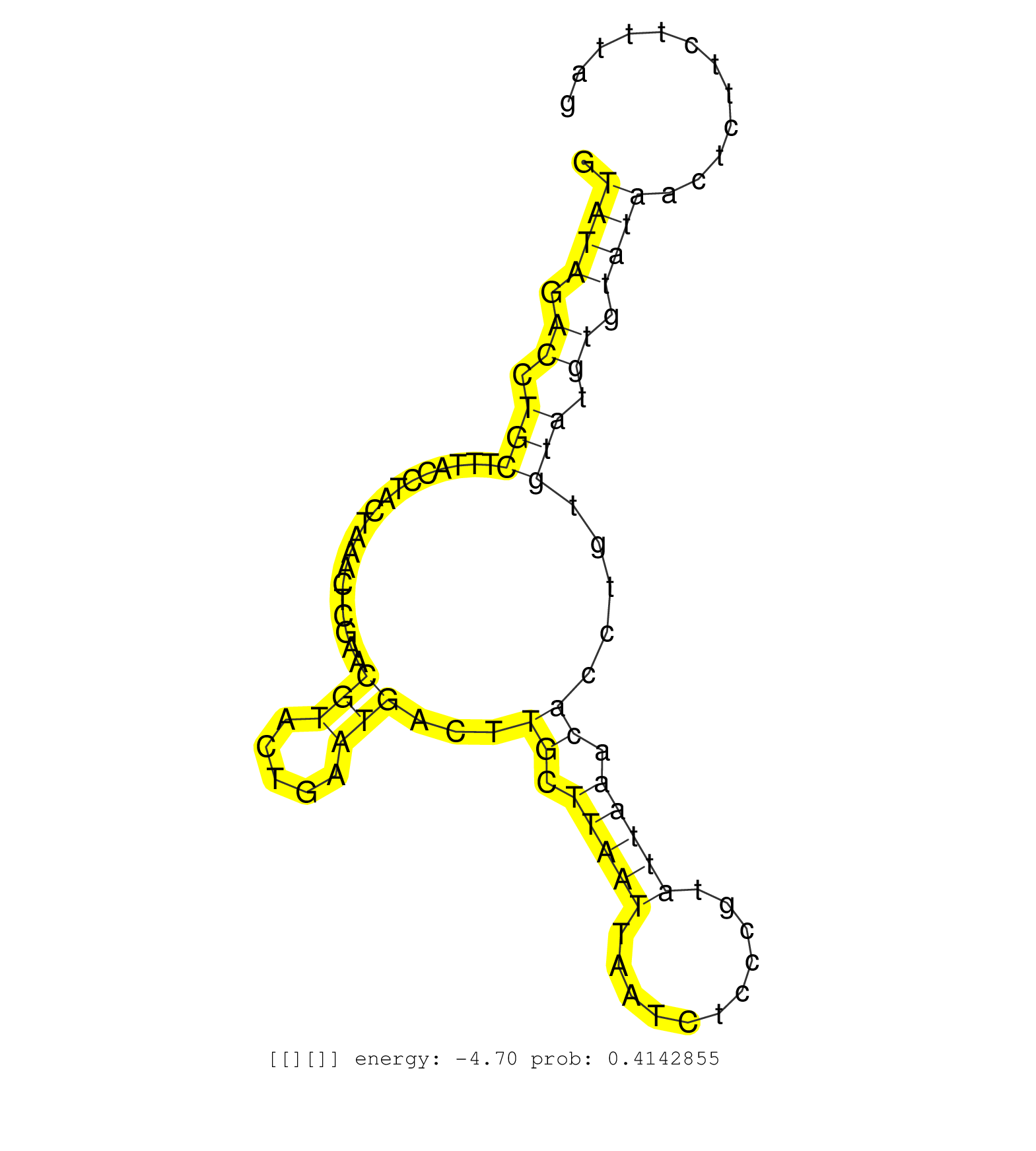

| Gene: Ubxd7 | ID: uc007yyq.1_intron_7_0_chr16_32381742_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| GATGTCACCTGAAAAGTCAGATGGAATAGTGGAGGGGATAGATGTAAATGGTGGGTTATGGTATAGACCTGCTTTACCTACTAAACTCGAACGTACTGAATGACTTGCTTAATTAATCTCCCGTATTAAACACCTGTGTATGTGTATAACTCTTCTTTAGGACCAAAAGCACAGCTGATGCTGCGGTACCCAGATGGAAAAAGGGAACAG .............................................................((((.((.(((...................(((.....)))...((.(((((...........))))).)).....))).)).)))).............................................................. ............................................................61.................................................................................................160................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGGGTTATGGTATAGACCTGCTTTACCTACTAAACTCGAACGTACTGAATG............................................................................................................ | 52 | 1 | 38.00 | 38.00 | 38.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........GAAAAGTCAGATGGAATAGTGGAGG............................................................................................................................................................................... | 25 | 1 | 5.00 | 5.00 | - | - | 3.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............AAGTCAGATGGAATAGTGGA................................................................................................................................................................................. | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..........GAAAAGTCAGATGGAATAGTGGAGGt.............................................................................................................................................................................. | 26 | t | 2.00 | 5.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TGGAATAGTGGAGGGGATAGATGTAAA.................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............AAAGTCAGATGGAATAGTGGAGG............................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...GTCACCTGAAAAGTCAGATGGAATAGTGG.................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........GAAAAGTCAGATGGAATAGTGGAGGa.............................................................................................................................................................................. | 26 | a | 1.00 | 5.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................CTTTAGGACCAAAAGCACAGCTGATGCTGC.......................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......CCTGAAAAGTCAGATGGAATAGTGGAGGGGATA.......................................................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................ATAGTGGAGGGGATAGATG...................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGATGCTGCGGTACCCAGATGGAAAAA........ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................ATGGAATAGTGGAGGGGATAGATGTAA................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGAAAAGTCAGATGGAATAGTGGAG................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGCTGCGGTACCCAGATGGAAAAAGGGAAt.. | 30 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TGGAATAGTGGAGGGGATAGATGTA.................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................ATAGTGGAGGGGATAGATGTAAATGGac............................................................................................................................................................. | 28 | ac | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TGCGGTACCCAGATGGAAAAAGGGAA... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................GGAATAGTGGAGGGGATAGATGTAAAT................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............GTCAGATGGAATAGTGGAGGGGATAGAT....................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................GAATAGTGGAGGGGATAGATGTAAATG................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGAAAAGTCAGATGGAATAGTGGAGG............................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................AGGGGATAGATGTAAATG................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................TGATGCTGCGGTACCCAGATGGAA........... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........GAAAAGTCAGATGGAATAGTGGAGGGGATAGAT....................................................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....TCACCTGAAAAGTCAGATGGAATAGTGG.................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| GATGTCACCTGAAAAGTCAGATGGAATAGTGGAGGGGATAGATGTAAATGGTGGGTTATGGTATAGACCTGCTTTACCTACTAAACTCGAACGTACTGAATGACTTGCTTAATTAATCTCCCGTATTAAACACCTGTGTATGTGTATAACTCTTCTTTAGGACCAAAAGCACAGCTGATGCTGCGGTACCCAGATGGAAAAAGGGAACAG .............................................................((((.((.(((...................(((.....)))...((.(((((...........))))).)).....))).)).)))).............................................................. ............................................................61.................................................................................................160................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|