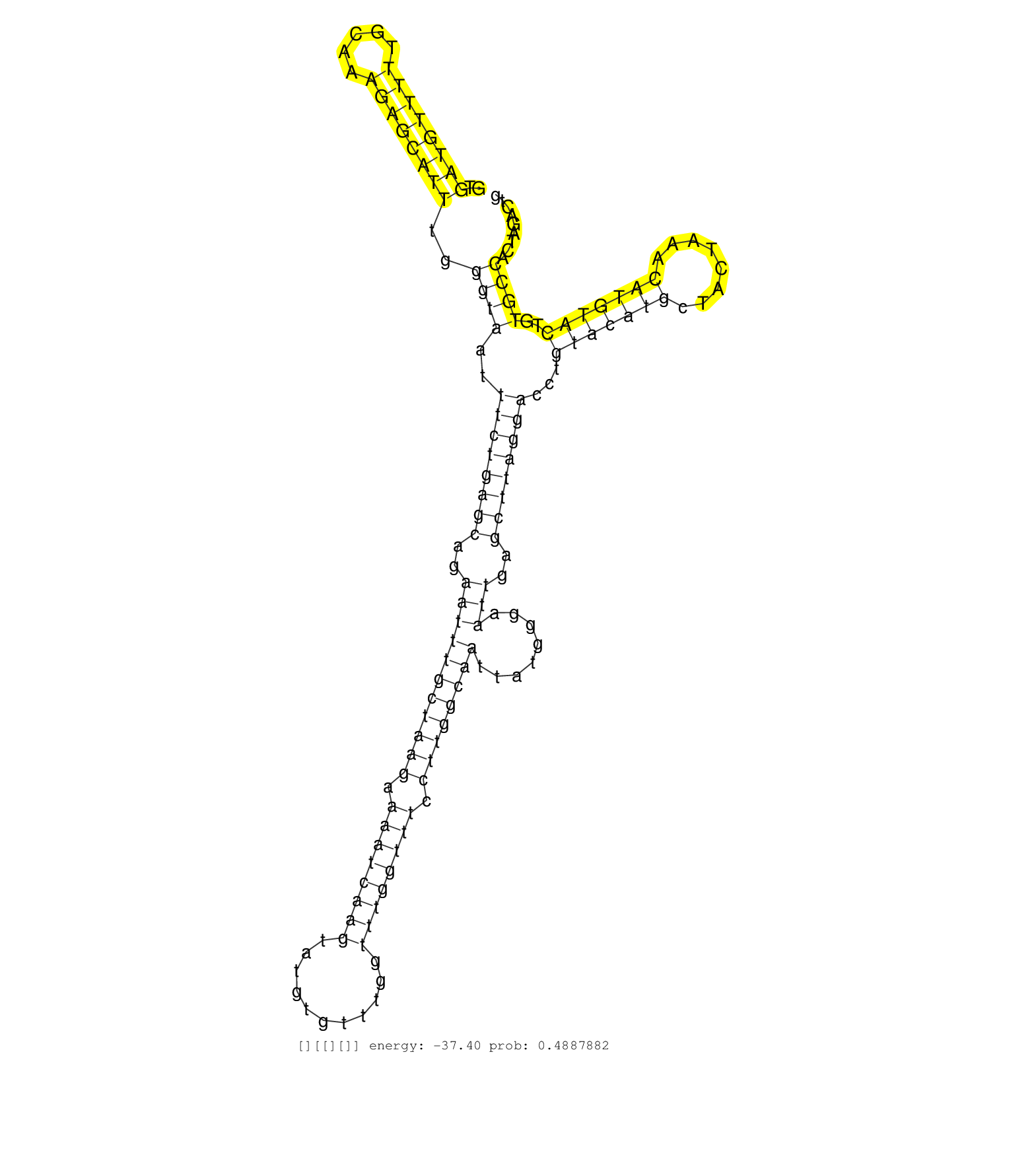

| Gene: Wdr53 | ID: uc007yym.1_intron_1_0_chr16_32252405_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(8) PIWI.ip |
(1) PIWI.mut |
(24) TESTES |
| TTCGACCTCAGAGGCCTCAGAGCCTGGTGTCATGTGGCCTGGATATGCAGGTGATGTTTTTGCAAAGAGCATTTGGGTAATTTCTGAGCAGAATTTGCTAAGAAAATCAAGTATGTGTTTGGTTTGGTTTCCTTGGCAATTATGGGAATTGAGCTTAGGACCTGTACATGCTACTAAACATGTACTGTGCCACTAGACTGTGTCCCCAACCCCTGGAATCAGATTATTGATCAGGTGTTTTCTACTCTCT ....................................................((((((((.....))))))))..((((..((((((((..(((((((((((.((((((((...........)))))))).))))))))........)))..))))))))...(((((((........)))))))..))))........................................................... ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................TACTAAACATGTACTGTGCCACTAGAC.................................................... | 27 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TGCAGGTGATGTTTTTGCAAAGAGCATT................................................................................................................................................................................. | 28 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TGCAGGTGATGTTTTTGCAAAGAGCATTT................................................................................................................................................................................ | 29 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................TGGCAATTATGGGAATTGAGCTTAGGACt........................................................................................ | 29 | t | 2.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TAAACATGTACTGTGCCACTAGACTG.................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TTATTGATCAGGTGTTTTCTACTCTC. | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TAAACATGTACTGTGCCACTAGACTGTGT............................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................AAAGAGCATTTGGGTAATTTCTGAGC................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................TGGCAATTATGGGAATTGAGCTTAGG........................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TAAGAAAATCAAGTATGTGTTTGGT............................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................TGCAAAGAGCATTTGGGTAATTTCTGAGC................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGATGTTTTTGCAAAGAGCATTTGGGTA........................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................ATCAAGTATGTGTTTGGTTTGGTTTC....................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGGAATCAGATTATTGATCAGGTGTT........... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TTGATCAGGTGTTTTCTACTCTCTaa | 26 | aa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TGTGTTTGGTTTGGTTTCCTTGGCA................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................ATGTGGCCTGGATATGCAGGTa..................................................................................................................................................................................................... | 22 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................TGGCCTGGATATGCAG........................................................................................................................................................................................................ | 16 | 2 | 1.00 | 1.00 | - | - | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................GGATATGCAGGTGATGTTTTTGCAA......................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................AGTATGTGTTTGGTTTGGTTTCCTTGGC................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........GAGGCCTCAGAGCCTGGTGTCATGTGGCCTGG................................................................................................................................................................................................................ | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................GAGCCTGGTGTCATGTGGCCTGGAT.............................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TAATTTCTGAGCAGAATTTGCTAAGAAAA................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................TGAGCAGAATTTGCTAAGA................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................TGGGAATTGAGCTTAGtgc......................................................................................... | 19 | tgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TACTAAACATGTACTGTGCCACTAGA..................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................AGCCTGGTGTCATGTGGCCT.................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TGGTGTCATGTGGCCTGGATATGC.......................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TAAACATGTACTGTGCCACTAGACTGTGTC.............................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TGTCATGTGGCCTGGATATGCAGGTGAT................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TTAGGACCTGTACATGC............................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TGTGGCCTGGATATGCAGGTGATG.................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................AGAGCATTTGGGTAATTTCTGAGCAG............................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGGCAATTATGGGAATTGAGCTTAGGAC......................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................GTGGCCTGGATATGCAG........................................................................................................................................................................................................ | 17 | 2 | 1.00 | 1.00 | - | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TCAGAGGCCTCAGAGCCTGGTGTC........................................................................................................................................................................................................................... | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGGGAATTGAGCTTAGGACC........................................................................................ | 20 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| TTCGACCTCAGAGGCCTCAGAGCCTGGTGTCATGTGGCCTGGATATGCAGGTGATGTTTTTGCAAAGAGCATTTGGGTAATTTCTGAGCAGAATTTGCTAAGAAAATCAAGTATGTGTTTGGTTTGGTTTCCTTGGCAATTATGGGAATTGAGCTTAGGACCTGTACATGCTACTAAACATGTACTGTGCCACTAGACTGTGTCCCCAACCCCTGGAATCAGATTATTGATCAGGTGTTTTCTACTCTCT ....................................................((((((((.....))))))))..((((..((((((((..(((((((((((.((((((((...........)))))))).))))))))........)))..))))))))...(((((((........)))))))..))))........................................................... ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................CCCAACCCCTGGAAacac................................ | 18 | acac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................................................................GACCTGTACATGCtttt............................................................................... | 17 | tttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |