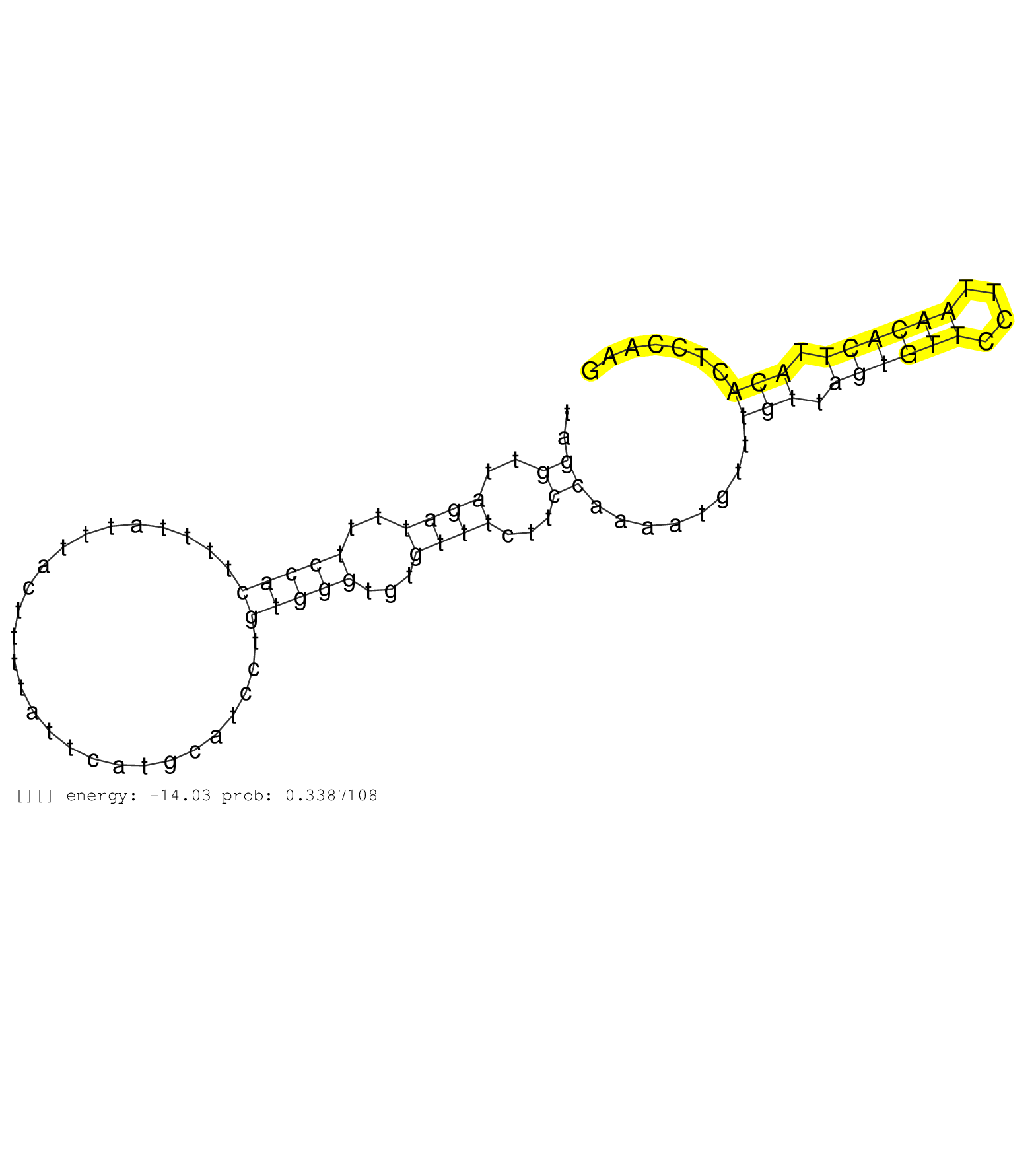
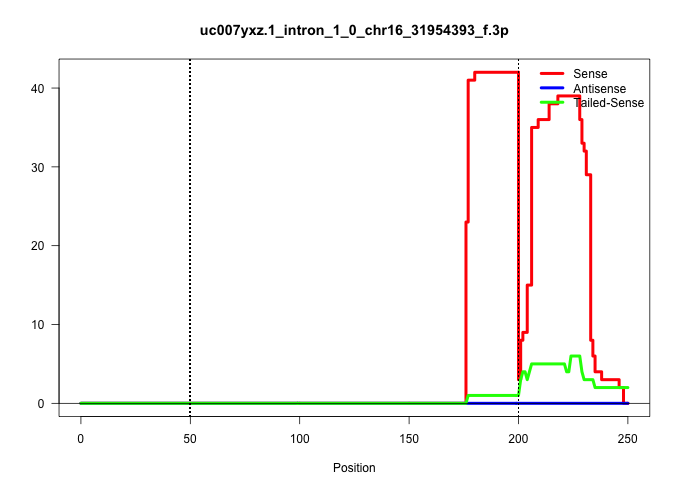
| Gene: Ncbp2 | ID: uc007yxz.1_intron_1_0_chr16_31954393_f.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(7) PIWI.ip |
(1) PIWI.mut |
(22) TESTES |
| TTACCTGTGCTTTACCTGTCCTTTACTGCCCAGAAGTCGTGTTAGACCAACTTTGAACTGACTTCTTCTACAAAATGAATCTCTATGTAAATAATCTGGTTAGGTTAGATTTTCCACTTTTATTTACTTTTATTCATGCATCCTGTGGGTGTGTTTCTTCCAAAATGTTTGTTAGTGTTCCTTAACACTTACACTCCAAGATACTATTCAAGAGCAGATGCGGAGAACGCAATGCGGTACATAAACGGAA ......................................................................................................((..((((..(((((...........................)))))...))))...))........(((.((((((....)))))).)))......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT1() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................GTTCCTTAACACTTACACTCCAAG.................................................. | 24 | 1 | 33.00 | 33.00 | 16.00 | 8.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | 3.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................................................TTCCTTAACACTTACACTCCAAG.................................................. | 23 | 1 | 21.00 | 21.00 | 7.00 | 4.00 | 5.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................................................................................................TTCAAGAGCAGATGCGGAGAACGCAAT................. | 27 | 1 | 17.00 | 17.00 | - | - | - | 3.00 | 5.00 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TATTCAAGAGCAGATGCGGAGAACGCA................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TACTATTCAAGAGCAGATGCGGAGAAC...................... | 27 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................................................................................................TACTATTCAAGAGCAGATGCGGAGAACG..................... | 28 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TATTCAAGAGCAGATGCGGAGAACGCAAT................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................TGCGGTACATAAACGG.. | 16 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TTCAAGAGCAGATGCGGAGAACGCAATG................ | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCCTTAACACTTACACTCCAAGA................................................. | 24 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TACTATTCAAGAGCAGATGCGGAGAACa..................... | 28 | a | 1.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CTTAACACTTACACTCCAAG.................................................. | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................AAGAGCAGATGCGGAGAACGCAAT................. | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TATTCAAGAGCAGATGCGGAGAACGCAA.................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................GAACGCAATGCGGTACATAAACGGgact | 28 | gact | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TACTATTCAAGAGCAGATGCGGAGAACGt.................... | 29 | t | 1.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TACTATTCAAGAGCAGATGCGGAGAACGC.................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TACTATTCAAGAGCAGATGCGGAGAACGCAA.................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................GAACGCAATGCGGTACATAAACGGAActaa | 30 | ctaa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCCTTAACACTTACACTCCAAGAaat.............................................. | 27 | aat | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................................................................................ATTCAAGAGCAGActta............................ | 17 | ctta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................ACTATTCAAGAGCAGATGCGGAGAACt..................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................ACTATTCAAGAGCAGATGCGGAGAACGCAAT................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGTTCCTTAACACTTACACTCCAAG.................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................CAGATGCGGAGAACGCAATGCGGT............ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TTCAAGAGCAGATGCGGAGAACGCAATGC............... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TTCAAGAGCAGATGCGGAGAACGCAATGt............... | 29 | t | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................GTTCCTTAACACTTACACTCCAAGA................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................CAGATGCGGAGAACGCAATGC............... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TGCGGAGAACGCAATGCGGTACATAAAC.... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| TTACCTGTGCTTTACCTGTCCTTTACTGCCCAGAAGTCGTGTTAGACCAACTTTGAACTGACTTCTTCTACAAAATGAATCTCTATGTAAATAATCTGGTTAGGTTAGATTTTCCACTTTTATTTACTTTTATTCATGCATCCTGTGGGTGTGTTTCTTCCAAAATGTTTGTTAGTGTTCCTTAACACTTACACTCCAAGATACTATTCAAGAGCAGATGCGGAGAACGCAATGCGGTACATAAACGGAA ......................................................................................................((..((((..(((((...........................)))))...))))...))........(((.((((((....)))))).)))......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT1() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|