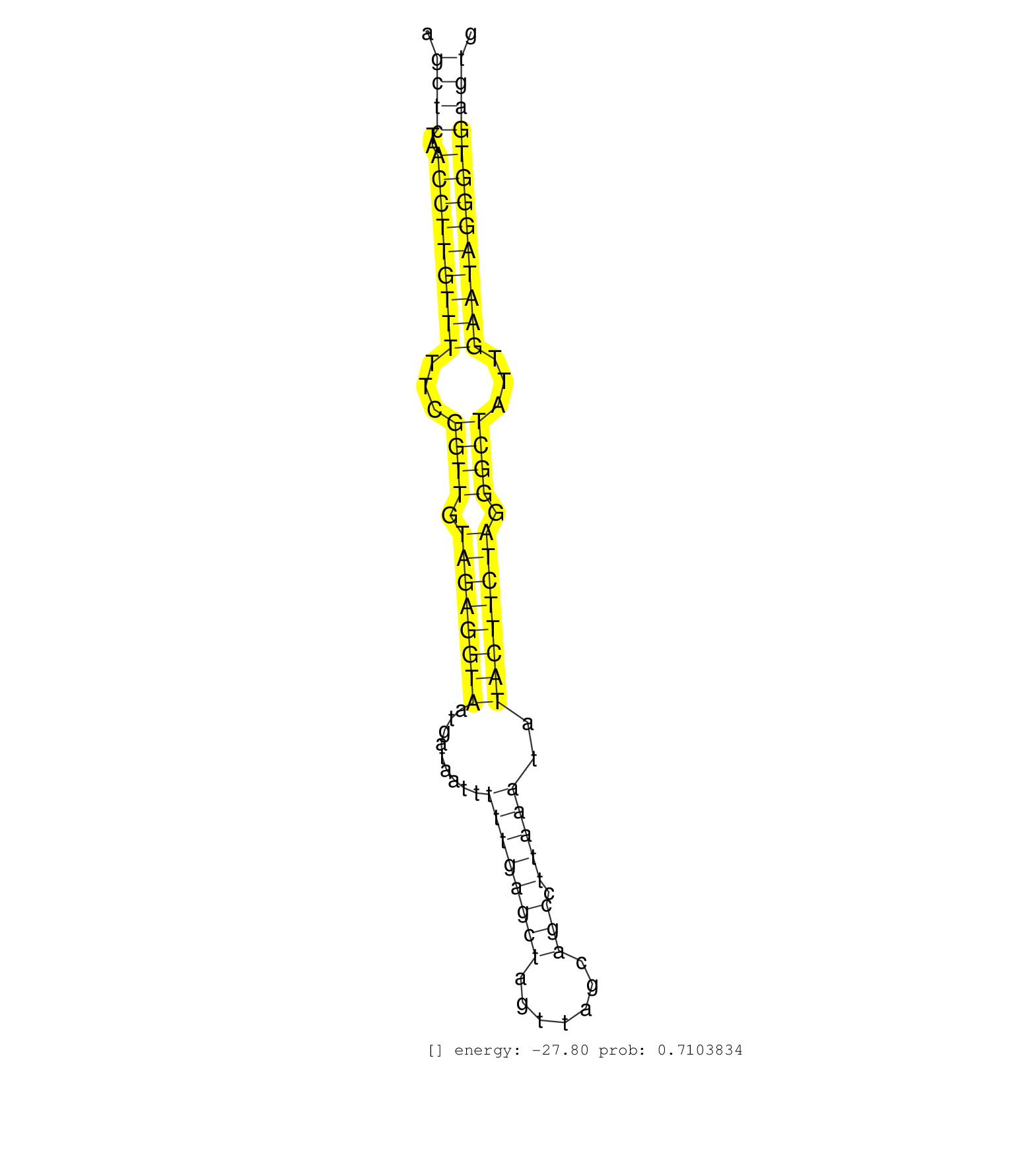

| Gene: Abcc5 | ID: uc007ypr.1_intron_5_0_chr16_20400113_r.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
| TGTCCATCGTGTGCCTGATGATCACGCAGTTGGCTGGCTTCAGTGGACCAGTAAGTTCTGACTATCCTTTCTGAACATCTCCAGACCCGGCCATGGCCAGCTCTAACCTTGTTTTTCGGTTGTAGAGGTAATGATAATTTTTGAGCTAGTTAGCAGCCTTAAATATACTTCTAGGGCTATTGAATAGGGTGAGTGGATTTGGGAGTGAACGGTGGCTTGTCAGCTAGACTGTTAAGAACAAAGAGCTGCA ...................................................................................................((((..(((((((((...((((.((((((((.........((((((((.......))).)))))..)))))))).))))...)))))))))))))........................................................ ..................................................................................................99..............................................................................................195..................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................TGATCACGCAGTTGGCTGGCTTCAGT.............................................................................................................................................................................................................. | 26 | 1 | 7.00 | 7.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 |
| ...............TGATGATCACGCAGTTGGCTGGCTTC................................................................................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TAGGGCTATTGAATAGGGTGAGTGGA..................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TACTTCTAGGGCTATTGAATAGGGTG........................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................TAACCTTGTTTTTCGGTTGTAGAGGTA........................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................GATCACGCAGTTGGCTGGCTTCAGTGGA........................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAACGGTGGCTTGTCAGCTA........................ | 21 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............CCTGATGATCACGCAGTTGGCT....................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TTTCTGAACATCTCCAGACCCGGCCATG........................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................ATGATCACGCAGTTGGCTGG..................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................ACTGTTAAGAACAAAGAGCTGCA | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TAGGGCTATTGAATAGGGTGAGTGG...................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TGATGATCACGCAGTTGGCTGGCTTCAGT.............................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CTAACCTTGTTTTTCGGTTGTAGAGGTA........................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................CTTGTCAGCTAGACTGTTAAGAACAAA........ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................ATCACGCAGTTGGCTGGCTTCAGTG............................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................ATATACTTCTAGGGCTATTGAATAGG.............................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TTCTGAACATCTCCAGACC................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TTCGGTTGTAGAGGTAATGATAATTTTT............................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................TGTTTTTCGGTTGTAGAGGTAATG..................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................TAGTTAGCAGCCTTAAATATACTTCTAG............................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................TGATCACGCAGTTGGCTGGCTTCAG............................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................ACCTTGTTTTTCGGTTGTAGAGGTA........................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................TATCCTTTCTGAACATCTCCAGACCCGGCC.............................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TATCCTTTCTGAACATCTCCAGACCCGGC............................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................CTGAACATCTCCAGACCCGGCCA............................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TCCTTTCTGAACATCTCCAGACCCGGC............................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................TTTCTGAACATCTCCAGACCCGGCCA............................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TGGCTTGTCAGCTAGACTGTTAAGAACA.......... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................................TACTTCTAGGGCTATTGAATA................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TAGGGCTATTGAATAGGGTGAGT........................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| TGTCCATCGTGTGCCTGATGATCACGCAGTTGGCTGGCTTCAGTGGACCAGTAAGTTCTGACTATCCTTTCTGAACATCTCCAGACCCGGCCATGGCCAGCTCTAACCTTGTTTTTCGGTTGTAGAGGTAATGATAATTTTTGAGCTAGTTAGCAGCCTTAAATATACTTCTAGGGCTATTGAATAGGGTGAGTGGATTTGGGAGTGAACGGTGGCTTGTCAGCTAGACTGTTAAGAACAAAGAGCTGCA ...................................................................................................((((..(((((((((...((((.((((((((.........((((((((.......))).)))))..)))))))).))))...)))))))))))))........................................................ ..................................................................................................99..............................................................................................195..................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|