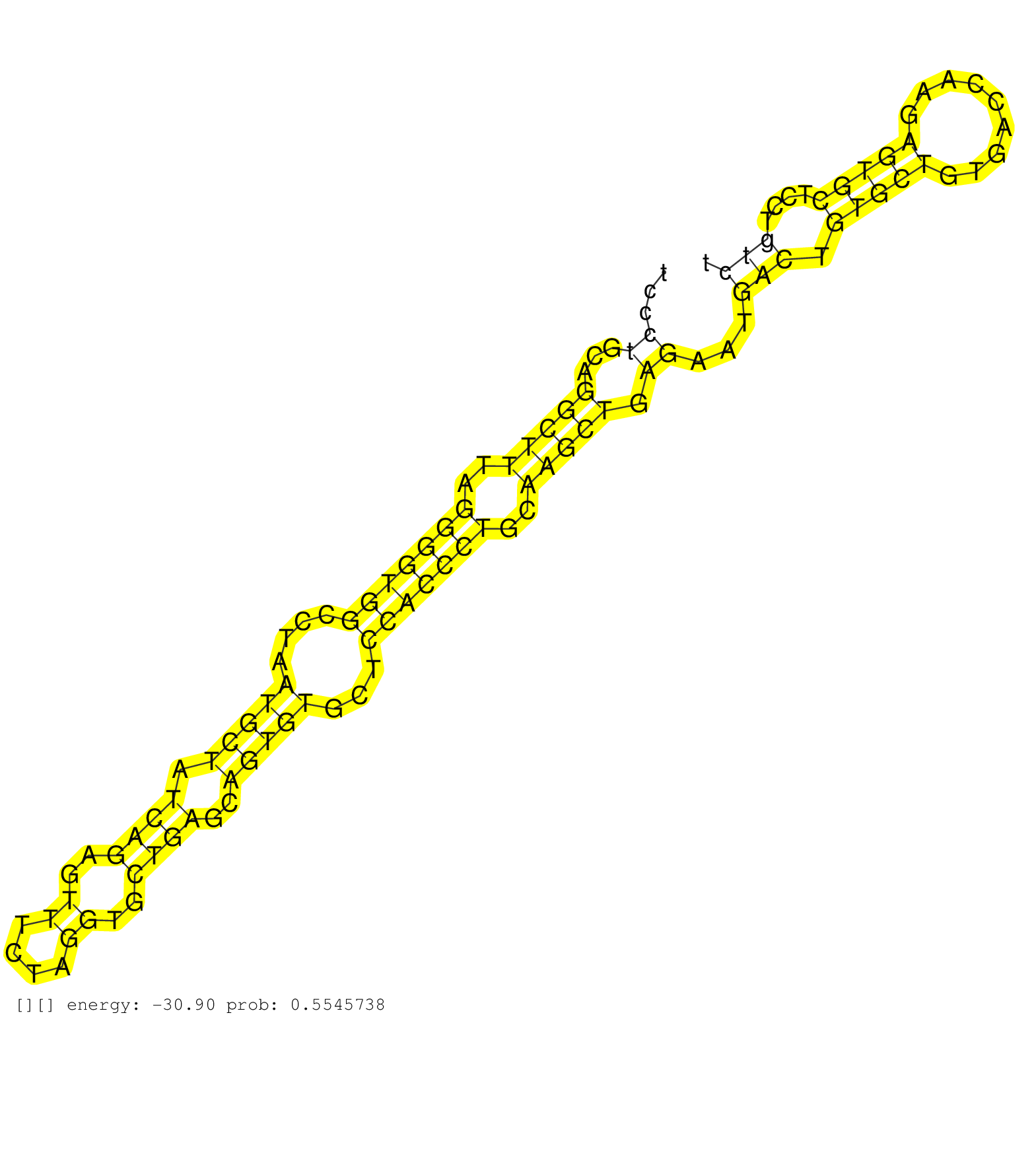

| Gene: Cdc45l | ID: uc007yol.1_intron_3_0_chr16_18785285_r.3p | SPECIES: mm9 |
 |
 |
|
(6) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| AGAGGGTTGAGCCAGTAGATAATTGGATGCTGAGAGCCAAAATATCAAGTTGGAATTTTAGCATACCCTCTAGTGACTGCATCCCTGCAGGCTTTAGGGGTGGCCTAATGCTATCAGAGTTTCTAGGTGCTGAGCAGTGTGCTCCACCCTGCAAGCTGAGAATGACTGTGCTGTGACCAAGAGTGCTCCTGTCTCCATAGACAAAGAATCGACGATGCAAGCTGCTGCCCCTGGTGATGGCTGCCCCGCT ....................................................................................((...(((((..(((((((....(((((.((((..((....))..))))..)))))...)))))))..))))).))...(((.(((((.........)))))....)))......................................................... .................................................................................82..............................................................................................................194...................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................GCAGGCTTTAGGGGTGGCCTAATGCTATCAGAGTTTCTAGGTGCTGAGCAGT................................................................................................................ | 52 | 1 | 28.00 | 28.00 | 28.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................AGAATCGACGATGCAAGCTGCTGCCCC................... | 27 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TCGACGATGCAAGCTGCTGCCCCTGGT............... | 27 | 1 | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................ACAAAGAATCGACGATGCAAGCTGCTt....................... | 27 | t | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................................................................................TAGACAAAGAATCGACGATGCAAGCTGC......................... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................AGAATCGACGATGCAAGCTGCTGCCCCT.................. | 28 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................ACTGTGCTGTGACCAAGAGTGCTCCT............................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................................GCAAGCTGAGAATGACTGTGCTGTGAC......................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................CAAAGAATCGACGATGCAAGCTGCTGCC..................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................AAGAATCGACGATGCAAGCTGCTGCCCC................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................AATCGACGATGCAAGCTGCTGCCCCcgg................ | 28 | cgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................................................................................................ACAAAGAATCGACGATGCAAGCTGCTGCCC.................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................................................................................................GACAAAGAATCGACGATGCAAGCTGCTGCCC.................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GACAAAGAATCGACGATGCAAGCTGCTGCC..................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................ATGACTGTGCTGTGACCAAGAG................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................CTGAGAATGACTGTGCTGTGACCAAGA.................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................................................................................................................ACAAAGAATCGACGATGCAAGCTGCTGC...................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GACAAAGAATCGACGATGCAAGCTGC......................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................AAAGAATCGACGATGCAAGCTGCTG....................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................................................................................................TAGACAAAGAATCGACGATGCAAGCTG.......................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................................................................................ACAAAGAATCGACGATGCAAGCTGCT........................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| AGAGGGTTGAGCCAGTAGATAATTGGATGCTGAGAGCCAAAATATCAAGTTGGAATTTTAGCATACCCTCTAGTGACTGCATCCCTGCAGGCTTTAGGGGTGGCCTAATGCTATCAGAGTTTCTAGGTGCTGAGCAGTGTGCTCCACCCTGCAAGCTGAGAATGACTGTGCTGTGACCAAGAGTGCTCCTGTCTCCATAGACAAAGAATCGACGATGCAAGCTGCTGCCCCTGGTGATGGCTGCCCCGCT ....................................................................................((...(((((..(((((((....(((((.((((..((....))..))))..)))))...)))))))..))))).))...(((.(((((.........)))))....)))......................................................... .................................................................................82..............................................................................................................194...................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................TGGATGCTGAGAGCCAAcgg.................................................................................................................................................................................................................. | 20 | cgg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |