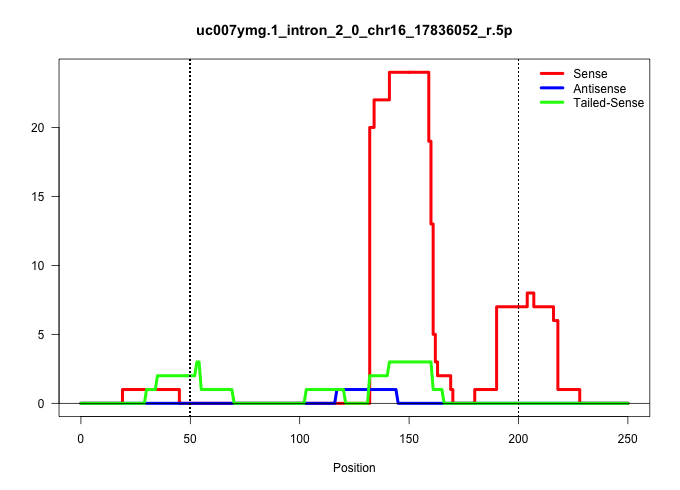
| Gene: Car15 | ID: uc007ymg.1_intron_2_0_chr16_17836052_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(6) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| ATGCACGGAGCCAACCAGATGGGTTTGCTATACTGGCTGTGCTGTTGGTGGTGAGGAAGGGGCTGTGGCCCCGTACATGTTTCCAGAAACAGGGACTTGGAGTTGTGTGGAGGGACCAGGGATCTTCAGCTCTGAAAGACTTAGAGAACTCAGAAGAGCTCACTGCCCCCCACCAACACATGCACACAAATGGGCTAAGCAGCTTTAACAATGGGATACACAGATGAGTCCAAATCCTCGTGAATGAAGA ...................................................................................................(((((...((((((.........)))))).((((((.....))))))..........)))))......................................................................................... ..................................................................................................99................................................................165................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................TGAAAGACTTAGAGAACTCAGAAGAGCTC......................................................................................... | 29 | 1 | 8.00 | 8.00 | - | 1.00 | 1.00 | 3.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGAAAGACTTAGAGAACTCAGAAGAGCT.......................................................................................... | 28 | 1 | 6.00 | 6.00 | 3.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................................................................................TGGGCTAAGCAGCTTTAACAATGGGATA................................ | 28 | 1 | 5.00 | 5.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGAAAGACTTAGAGAACTCAGAAGAGC........................................................................................... | 27 | 1 | 5.00 | 5.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGAAAGACTTAGAGAACTCAGAAGAGCTt......................................................................................... | 29 | t | 2.00 | 6.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................CTGGCTGTGCTGTTGGTG........................................................................................................................................................................................................ | 18 | 2 | 1.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | 1.50 | - | - | - | - | - |
| ....................................................................................................................................................................................TGCACACAAATGGGCTAAGCAGCTTTA........................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TGTGTGGAGGGACCctaa................................................................................................................................. | 18 | ctaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................................TTAACAATGGGATACACAGATGAG...................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TAGAGAACTCAGAAGAGCTCACTGt.................................................................................... | 25 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................TACTGGCTGTGCTGTTGGTGGagga................................................................................................................................................................................................... | 25 | agga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................TAGAGAACTCAGAAGAGCTCACTGCCCCC................................................................................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TAGAGAACTCAGAAGAGCTCACTGCCCC................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................AAAGACTTAGAGAACTCAGAAGAGCTCA........................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGGAAGGGGCTGTGttt.................................................................................................................................................................................... | 17 | ttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................GCTGTGCTGTTGGTGGTata................................................................................................................................................................................................... | 20 | ata | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................AAAGACTTAGAGAACTCAGAAGAGCTCAC....................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................TGGGTTTGCTATACTGGCTGTGCTGT............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TGGGCTAAGCAGCTTTAACAATGGGA.................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGAAAGACTTAGAGAACTCAGAAGAGCTCA........................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ATGCACGGAGCCAACCAGATGGGTTTGCTATACTGGCTGTGCTGTTGGTGGTGAGGAAGGGGCTGTGGCCCCGTACATGTTTCCAGAAACAGGGACTTGGAGTTGTGTGGAGGGACCAGGGATCTTCAGCTCTGAAAGACTTAGAGAACTCAGAAGAGCTCACTGCCCCCCACCAACACATGCACACAAATGGGCTAAGCAGCTTTAACAATGGGATACACAGATGAGTCCAAATCCTCGTGAATGAAGA ...................................................................................................(((((...((((((.........)))))).((((((.....))))))..........)))))......................................................................................... ..................................................................................................99................................................................165................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................TCTTCAGCTCTGAAaccc.................................................................................................................. | 18 | accc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................AGGGATCTTCAGCTCTGAAAGACTTAGA......................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................GTCCAAATCCTCgtgg........... | 16 | gtgg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |