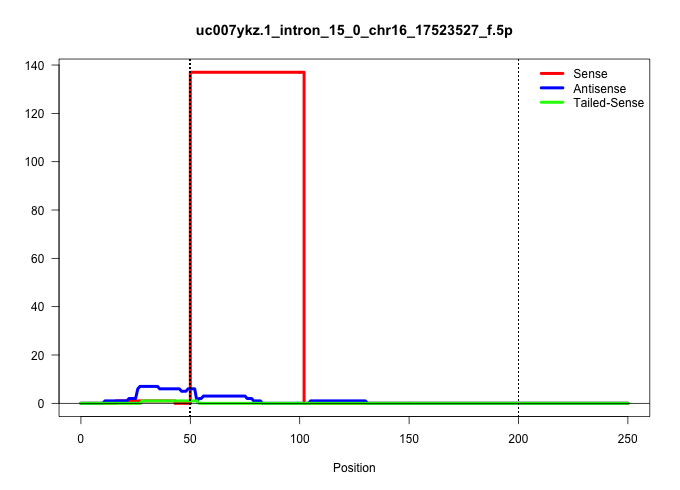
| Gene: Lztr1 | ID: uc007ykz.1_intron_15_0_chr16_17523527_f.5p | SPECIES: mm9 |
 |
|
(4) PIWI.ip |
(10) TESTES |
| GAAGCAGCAGCCGCCCCCTCGCACACCCTCCGATCAGCCTGTGGACATTGGTAGGGAACCTCTCTTCCTGTCCTGAGTATAGGGAAGTGGTAAGAGACTTAGATCCACTTACCTGCCAGTCCCTTGAGGTAAGGGTGGCTCTGTGACCCACTCAGGTGCTCCCTGGCACAGGGACTGGGAAGGACAGGGGCTTTTAATAGTAAGCTATGTGCCAGCAAGCCTGGGTACCCATGAAACCCACTCTTGAGGG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGGAACCTCTCTTCCTGTCCTGAGTATAGGGAAGTGGTAAGAGACTTAG.................................................................................................................................................... | 52 | 1 | 137.00 | 137.00 | 137.00 | - | - | - | - | - | - | - | - | - |
| ............................TCCGATCAGCCTGTGGACATTGGcac.................................................................................................................................................................................................... | 26 | cac | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ................CCTCGCACACCCTCCGATCAGCCTGTG............................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| GAAGCAGCAGCCGCCCCCTCGCACACCCTCCGATCAGCCTGTGGACATTGGTAGGGAACCTCTCTTCCTGTCCTGAGTATAGGGAAGTGGTAAGAGACTTAGATCCACTTACCTGCCAGTCCCTTGAGGTAAGGGTGGCTCTGTGACCCACTCAGGTGCTCCCTGGCACAGGGACTGGGAAGGACAGGGGCTTTTAATAGTAAGCTATGTGCCAGCAAGCCTGGGTACCCATGAAACCCACTCTTGAGGG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................CCTCCGATCAGCCTGTGGACATTGGTA..................................................................................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | - | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - |
| ...............................................................CTGTCCTGAGTATAagca......................................................................................................................................................................... | 18 | agca | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................AACCTCTCTTCCTGTCCTGAGTATAGG....................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................GGTAGGGAACCTCTCTTCCTGTCCTGA.............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................CACTTACCTGCCAGTCCCTTGAGGTA....................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................ACACCCTCCGATCAGCCTGTGGAC............................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................CTCCGATCAGCCTGTGGACATTGGTA..................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................GGAACCTCTCTTCCTGTCCTGAGTA........................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........CGCCCCCTCGCACACCCTCCGATCA...................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |