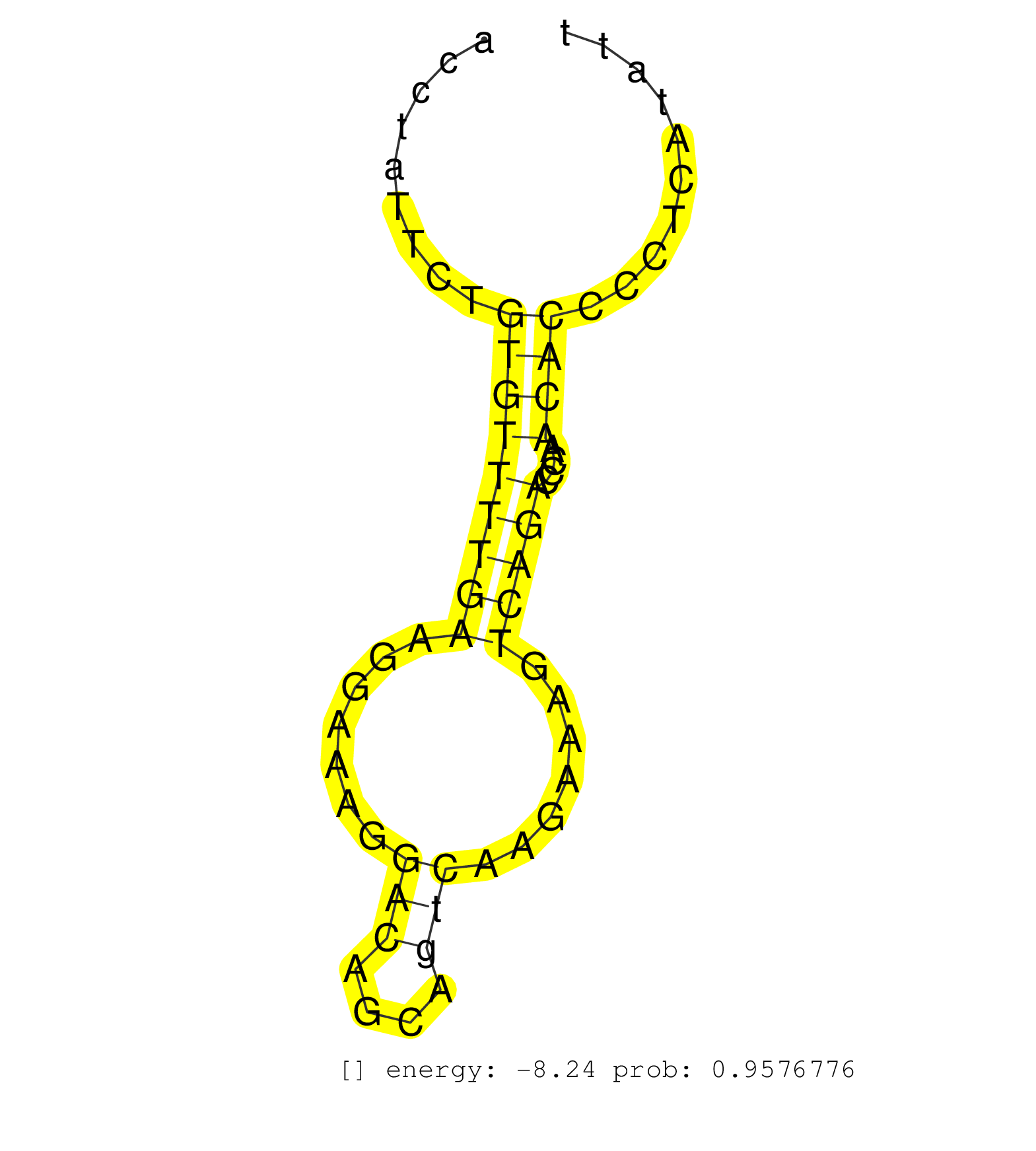

| Gene: Top3b | ID: uc007yjj.1_intron_16_0_chr16_16890776_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| GTGAGGGAAGAAGAAATGGGGAAGTATGGTACCTGGGCTCATACCAGCGGTTGAGGTATTTGCCTGGCACTGAATTCTGTATTCTGTGTACCTGTTAGGTGTTTAGCATTACCTATTCTGTGTTTTGAAGGAAAGGACAGCAGTCAAGAAAGTCAGACCCAACACCCCTCATATTCCTTCTAATTGCCCCCATCCCATAGGCCAAGCCAAGTCGCCTGCACTGCTCCCACTGTGATGAGACCTACACCCT .......................................................................................................................(((((((((.......(((....))).......)))))....))))..................................................................................... ..............................................................................................................111.............................................................175......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................TTCTGTGTTTTGAAGGAAAGGACAGCA............................................................................................................ | 27 | 1 | 27.00 | 27.00 | 9.00 | 12.00 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - |
| ...................................................................................................................TTCTGTGTTTTGAAGGAAAGGACAGC............................................................................................................. | 26 | 1 | 21.00 | 21.00 | 11.00 | 5.00 | 2.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................TTGAAGGAAAGGACAGCAGTCAAGAAA................................................................................................... | 27 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCTGTATTCTGTGTACCTGTTAGGTGT.................................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TTTGAAGGAAAGGACAGCAGTCAAGAA.................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TGGTACCTGGGCTCATACCAGCGGTT...................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................TGAAGGAAAGGACAGCAGTCAAGAAAGT................................................................................................. | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TAGGCCAAGCCAAGTCGCCTGCACTGCTC........................ | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TATTCTGTGTTTTGAAGGAAAGGACAGCA............................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................AGGAAAGGACAGCAGTCAAGAAAGTCAG.............................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCTAATTGCCCCCATCCCATAGGCCAAGCC.......................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................TGTGTTTTGAAGGAAAGGACAGCAGT.......................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGTTTTGAAGGAAAGGACAGCAGTCAA....................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TGAAGGAAAGGACAGCAGTCAAGAA.................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................TACCAGCGGTTGAGGTATTTGCCTGG....................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGTATTCTGTGTACCTGTTAGGTGT.................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................TGAAGGAAAGGACAGCAGTCAAGAAAaa................................................................................................. | 28 | aa | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TCTGTATTCTGTGTACCTGTTAGGTG..................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................TTCTGTGTACCTGTTAGGTGTTTAGC............................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TATTCTGTGTTTTGAAGGAAAGGACAG.............................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TCCCATAGGCCAAGCCAAGTCGCCcgc............................... | 27 | cgc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TTGAAGGAAAGGACAGCAGTCAAGAA.................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................AGCCAAGTCGCCTGCACTGCTCCCA..................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................ATTCTGTGTTTTGAAGGAAAGGACAGCA............................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TTCTGTGTTTTGAAGGAAAGGACAGCAG........................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGTTTTGAAGGAAAGGACAGCAGTCAAG...................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TATTCTGTGTTTTGAAGGAAAGGACAGC............................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................TGAATTCTGTATTCTGTGTACCTGTT.......................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TGAAGGAAAGGACAGCAGTCAAGAAAGTC................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TGAAGGAAAGGACAGCAGTCAAGAAA................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TCCCACTGTGATGAGACCTACACC.. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................................................................................CAAGAAAGTCAGACCCAACACCCCTCA............................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| GTGAGGGAAGAAGAAATGGGGAAGTATGGTACCTGGGCTCATACCAGCGGTTGAGGTATTTGCCTGGCACTGAATTCTGTATTCTGTGTACCTGTTAGGTGTTTAGCATTACCTATTCTGTGTTTTGAAGGAAAGGACAGCAGTCAAGAAAGTCAGACCCAACACCCCTCATATTCCTTCTAATTGCCCCCATCCCATAGGCCAAGCCAAGTCGCCTGCACTGCTCCCACTGTGATGAGACCTACACCCT .......................................................................................................................(((((((((.......(((....))).......)))))....))))..................................................................................... ..............................................................................................................111.............................................................175......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|