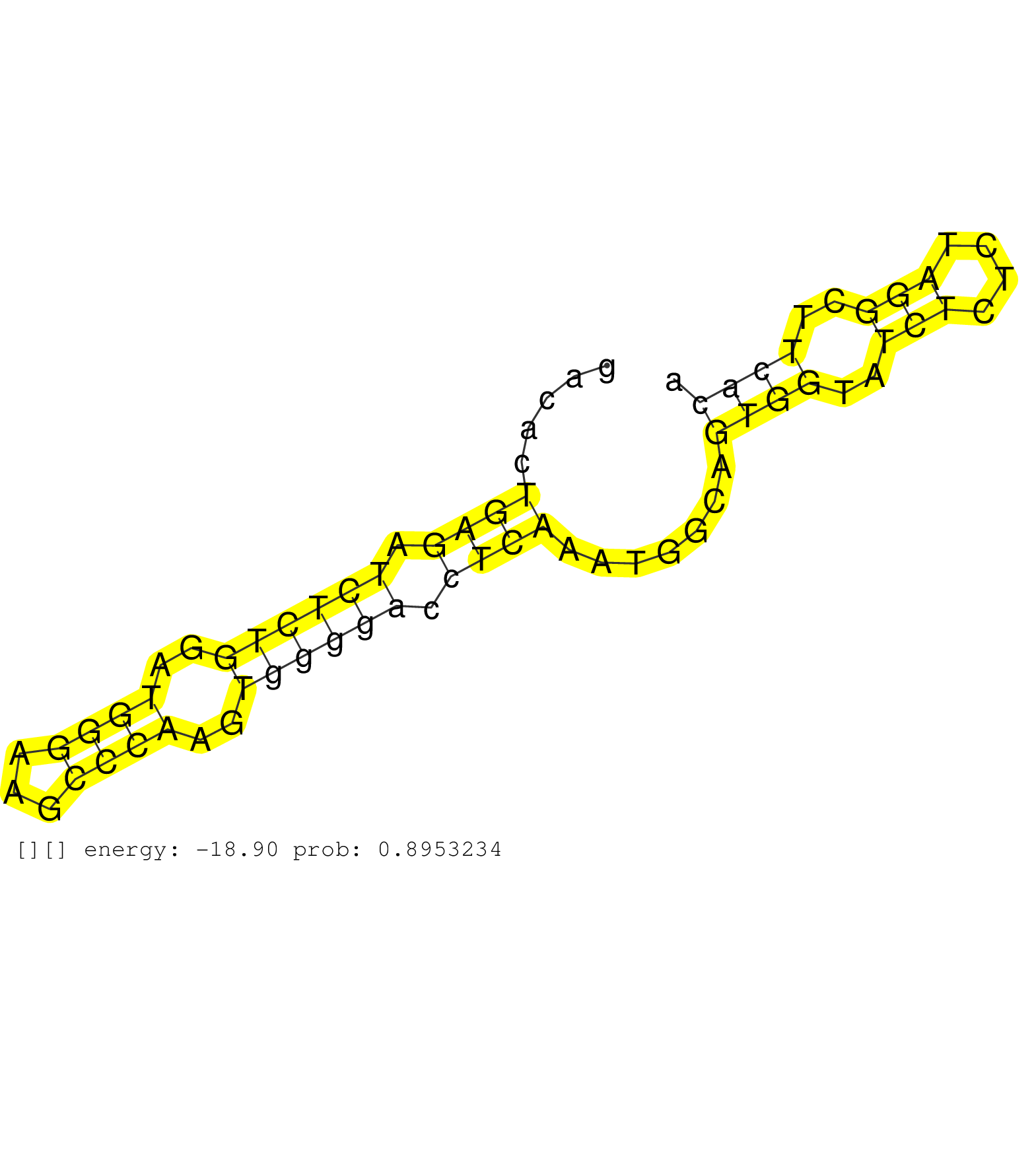

| Gene: Top3b | ID: uc007yjj.1_intron_12_0_chr16_16887817_f | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(3) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(19) TESTES |
| GATCGGGCCTGAACACTTTACATGCATGGGAAAAACAGTTATTTCTCCAGGTACAGTGGCAGACCACGGGAGTCCCTTTCTTCTCATCTATACAGACACTGAGATCTCTGGATGGGAAGCCCAAGTGGGGACCTCAAATGGCAGTGGTATCTCTCTAGGCTTCACAGAGATCATGCCTTGGCAGAGCGTGCCCCTGGAAGAGAGTCTG ...................................................................................................((((.((((((..((((...))))..)))))).)))).......((((..(((....)))..))))........................................... ..............................................................................................95.....................................................................166........................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................TGAGATCTCTGGATGGGAAGCCCAAGT.................................................................................. | 27 | 1 | 19.00 | 19.00 | 8.00 | 4.00 | - | 3.00 | - | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TACAGACACTGAGATCTCTGGATGG............................................................................................. | 25 | 1 | 5.00 | 5.00 | - | - | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TGAGATCTCTGGATGGGAAGCCCAAG................................................................................... | 26 | 1 | 5.00 | 5.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TACAGACACTGAGATCTCTGGATGGGA........................................................................................... | 27 | 1 | 4.00 | 4.00 | 1.00 | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TCAAATGGCAGTGGTATCTCTCTAGGCTT.............................................. | 29 | 1 | 3.00 | 3.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TCAAATGGCAGTGGTATCTCTCTAGGC................................................ | 27 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TACAGACACTGAGATCTCTGGATGGG............................................................................................ | 26 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...CGGGCCTGAACACTTTACATGCATGGa.................................................................................................................................................................................. | 27 | a | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 |
| .....................................................................................................................................TCAAATGGCAGTGGTATCTCTCTAGG................................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................CAGACACTGAGATCTCTGGATGGGAAG......................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CACTGAGATCTCTGGATGGGAAGCCC...................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGAACACTTTACATGCATGGGAAAAAC............................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................................................................................TGCCTTGGCAGAGCGTGCCCCTGGAAG........ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TACAGACACTGAGATCTCTGGATGGGt........................................................................................... | 27 | t | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...CGGGCCTGAACACTTTACATGCATGGGAA................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TATTTCTCCAGGTACAGTGGCAGACC............................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............CACTTTACATGCATGGGAAAAACAGTT........................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...CGGGCCTGAACACTTTACATGCATGGGA................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................TACAGACACTGAGATCTCTGGATGGGAAGC........................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..TCGGGCCTGAACACTTTACATGCATGG................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................AGACACTGAGATCTCTGGATGGGAAGC........................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................ACAGACACTGAGATCTCTGGATGGGAAGC........................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CAGAGATCATGCCTTGGCAGAGCGTGt................. | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TACAGACACTGAGATCTCTGGATG.............................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGGCAGAGCGTGCCCCTGGAAGA....... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................CAGACACTGAGATCTCTGGATGGGAAGC........................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................GGACCTCAAATGGCAGTGGTATCTCTCT.................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGAACACTTTACATGCATGGGAAAA.............................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TACATGCATGGGAAAAACAGTTATTTC................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTACAGTGGCAGACCACGGGAGTCCCTTT................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................CTATACAGACACTGAGATCTCTGGATGG............................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TCATCTATACAGACACTGAGATCTCTGGA................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TACATGCATGGGAAAAACAGTTATTTCTCC................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................TACATGCATGGGAAAAACAGTTATTTCTCCAGt............................................................................................................................................................. | 33 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................CACAGAGATCATGCCTTGGCAGAGCGT................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..TCGGGCCTGAACACTTTACATGCATGGG.................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TCGGGCCTGAACACTTTACATGCATGGGA................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GATCGGGCCTGAACACTTTACATGCATGGGAAAAACAGTTATTTCTCCAGGTACAGTGGCAGACCACGGGAGTCCCTTTCTTCTCATCTATACAGACACTGAGATCTCTGGATGGGAAGCCCAAGTGGGGACCTCAAATGGCAGTGGTATCTCTCTAGGCTTCACAGAGATCATGCCTTGGCAGAGCGTGCCCCTGGAAGAGAGTCTG ...................................................................................................((((.((((((..((((...))))..)))))).)))).......((((..(((....)))..))))........................................... ..............................................................................................95.....................................................................166........................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................AGGCTTCACAGAGAtgtt...................................... | 18 | tgtt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |