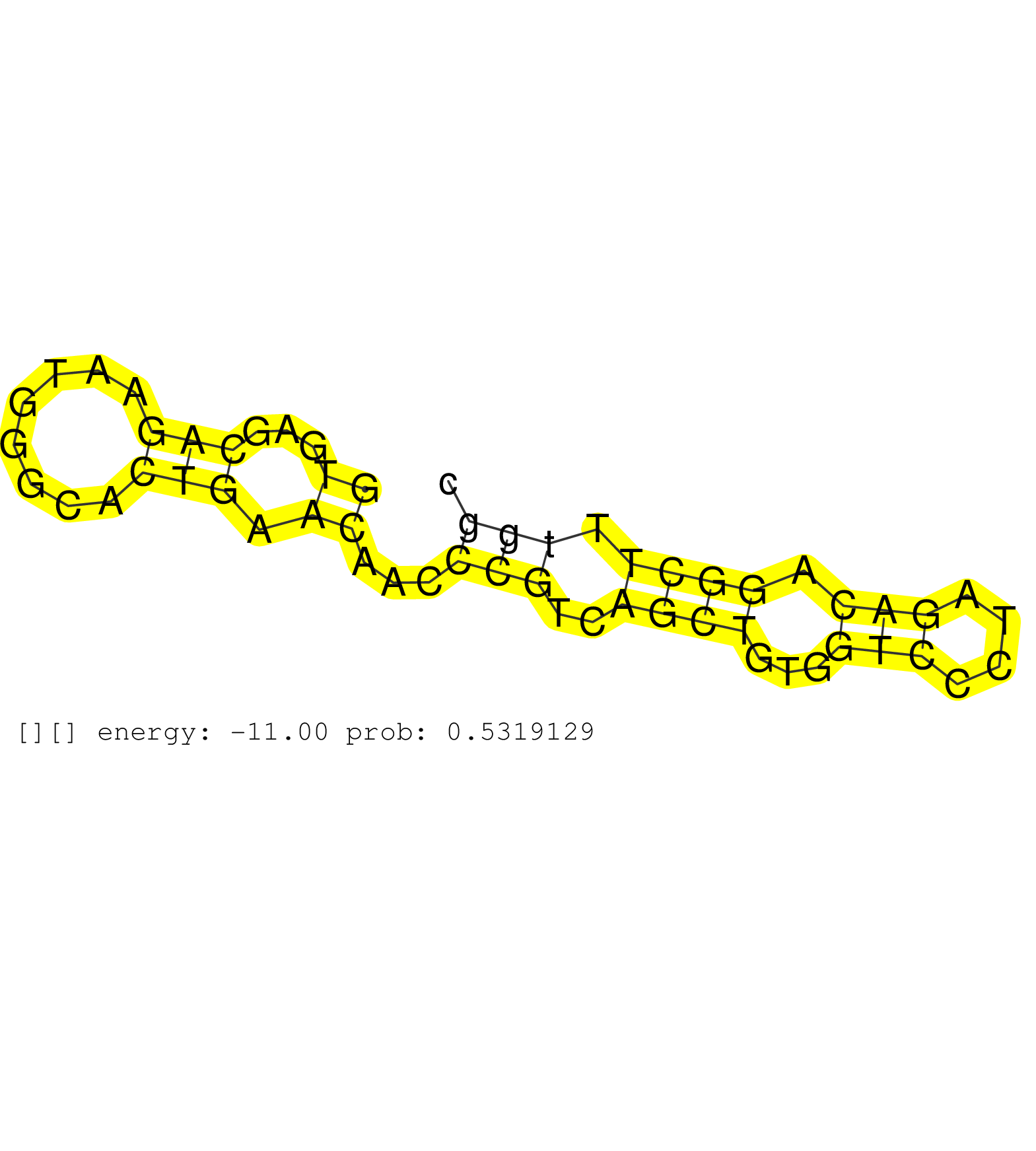

| Gene: Top3b | ID: uc007yjj.1_intron_10_0_chr16_16886853_f | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(15) TESTES |
| TGAAGGGCTCTTTGCGACAGCAGGCCAATCACCCATACTGGGCAGACTCGGTGAGCAGAATGGGCACTGAACAACCCGTCAGCTGTGGTCCCTAGACAGGCTTTGGCCAAACCCAAAACTGATGGAAGCCATCAGCCTGACTTGACTCCGCTTTGCCTCCCCTTCCTCAGGTAAAGCAGTTGTTAGCGGAAGGCATCAACCGCCCTCGGAAAGGTCATGA ..................................................((...(((........))).))...(((..((((...(((....))).)))).))).................................................................................................................. ..................................................51......................................................107............................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................TACTGGGCAGACTCGGTGAGCAGAATG.............................................................................................................................................................. | 27 | 1 | 6.00 | 6.00 | 3.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCAGAATGGGCACTGAACAACCCGT............................................................................................................................................. | 29 | 1 | 5.00 | 5.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - |
| ...................................TACTGGGCAGACTCGGTGAGCAGAAT............................................................................................................................................................... | 26 | 1 | 5.00 | 5.00 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................TACTGGGCAGACTCGGTGAGCAGAATGG............................................................................................................................................................. | 28 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................AATGGGCACTGAACAACCCGTCAGCTGT...................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGCAGAATGGGCACTGAACAACCCGT............................................................................................................................................. | 28 | 1 | 3.00 | 3.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCAGAATGGGCACTGAACAACCCtt............................................................................................................................................. | 29 | tt | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................CCGTCAGCTGTGGTCCCTAGACAGGCTT..................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TGGGCAGACTCGGTGAGCAGAATGGGCAC......................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCAGAATGGGCACTGAACAACCC............................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................TGGGCAGACTCGGTGAGCAGAATGGG............................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TTCCTCAGGTAAAGCAGTTGTTAGCGG............................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................................TCCTCAGGTAAAGCAGTTGTTAGCGGAA............................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCAGAATGGGCACTGAACAACCCG.............................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................GGCAGACTCGGTGAGCAGAATGGGCAC......................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TCAGCCTGACTTGACTCCGCTTTGCCTCC............................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................CTGGGCAGACTCGGTGAGCAGAATGGtt........................................................................................................................................................... | 28 | tt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................TACTGGGCAGACTCGGTGAGCAGAATa.............................................................................................................................................................. | 27 | a | 1.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................CCCGTCAGCTGTGGTCCCTAGACAGGCT...................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCAGAATGGGCACTGAACAACC................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................TGGGCACTGAACAACCCGTCAGCTGTGG.................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................AAGCAGTTGTTAGCGGAAGGCATCAACC................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................ACTGGGCAGACTCGGTGAGCAGAATG.............................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................CCCGTCAGCTGTGGTCCCTAGACAGGC....................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCAGAATGGGCACTGAACAAC................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................ACCCGTCAGCTGTGGTCCCTAGACAGG........................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGAAGGGCTCTTTGCGACAGCAGGCCAATCACCCATACTGGGCAGACTCGGTGAGCAGAATGGGCACTGAACAACCCGTCAGCTGTGGTCCCTAGACAGGCTTTGGCCAAACCCAAAACTGATGGAAGCCATCAGCCTGACTTGACTCCGCTTTGCCTCCCCTTCCTCAGGTAAAGCAGTTGTTAGCGGAAGGCATCAACCGCCCTCGGAAAGGTCATGA ..................................................((...(((........))).))...(((..((((...(((....))).)))).))).................................................................................................................. ..................................................51......................................................107............................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................CAAAACTGATGGAAatgt............................................................................................. | 18 | atgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................GCACTGAACAACCCG.............................................................................................................................................. | 15 | 3 | 0.33 | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - |