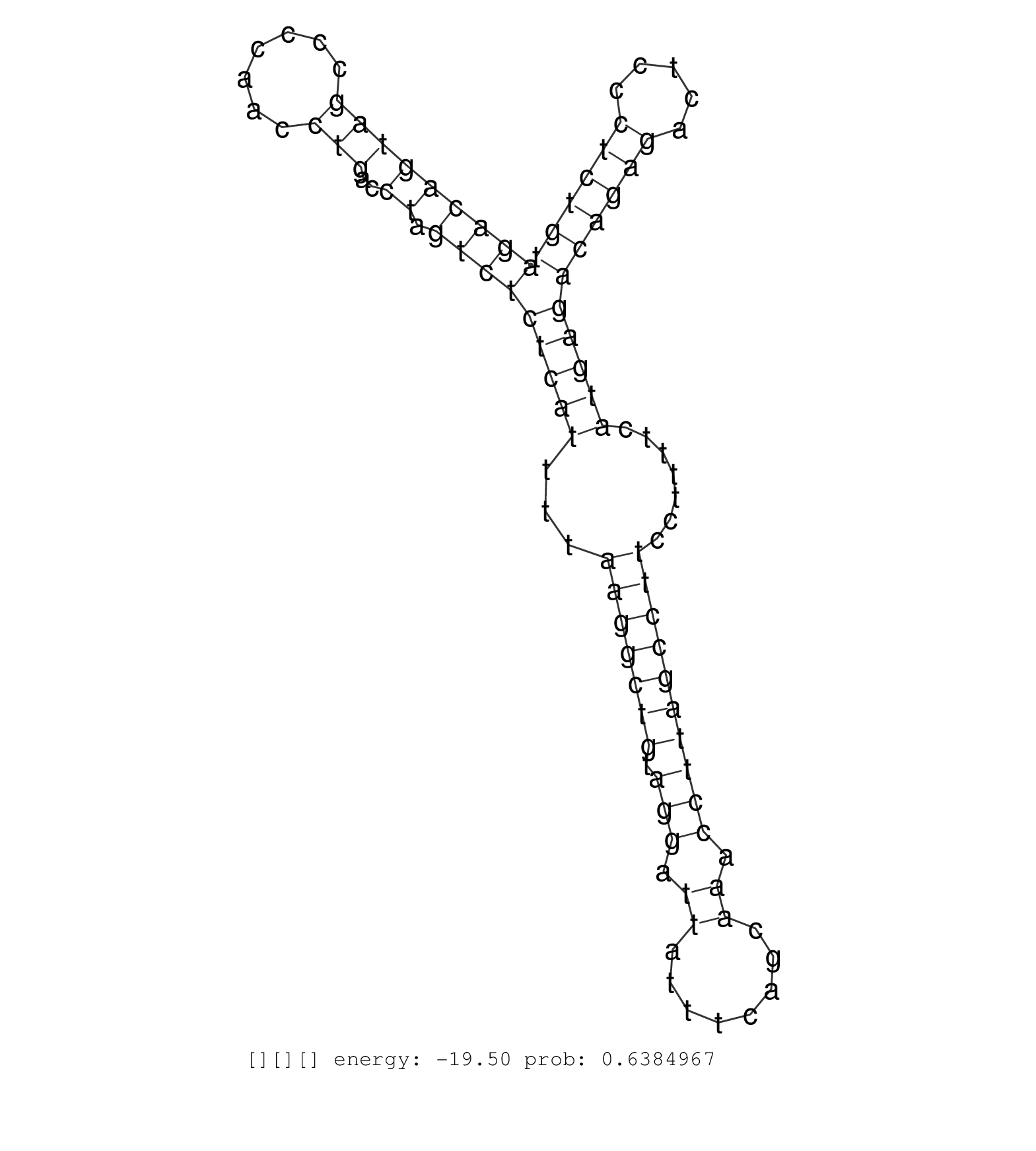

| Gene: Nde1 | ID: uc007ygy.1_intron_7_0_chr16_14190326_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(6) PIWI.ip |
(1) TDRD1.ip |
(19) TESTES |
| AGAGATGGTGTTGACAGAAGGCCAGGCAGTACCAGTGTGGGCGATAAAGGGTCAGTACCTTTGATACTCTCTGTCAGGACATACCAGGCTGCTCTAGATCAGACAGTAGCCCCAACCTGACCTAGTCTCTCATTTTAAGGCTGTAGGATTATTTCAGCAAACCTTAGCCTTCCTTTTCATGAGACAGAGACTCCCTCTGTAGCCCAGGCTGGCCTTGAATAAGCTTCTCGCTTGAGGGCAAATACGATGA ....................................................................................................(((((((((.......)))..)).))))(((((...(((((((.(((.((........)).)))))))))).......)))))((((((.....)))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................TGATACTCTCTGTCAGGACATACCAGGC................................................................................................................................................................. | 28 | 1 | 7.00 | 7.00 | 2.00 | 1.00 | - | - | - | 1.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................AGTGTGGGCGATAAAGG........................................................................................................................................................................................................ | 17 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - |
| ................................................................TACTCTCTGTCAGGACATACCAGGCT................................................................................................................................................................ | 26 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................TTGATACTCTCTGTCAGGACATACCAGGC................................................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TAGATCAGACAGTAGCCCCAACCTGAC................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TTGATACTCTCTGTCAGGACATACCAGG.................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGGCCTTGAATAAGCTTCTCGCTTG................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................CAGTGTGGGCGATAAAGG........................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TGTAGCCCAGGCTGGCCTTGAATAAa........................... | 26 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TACTCTCTGTCAGGACATACCAGGC................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........TGTTGACAGAAGGCCAGGCAGTACCAGT...................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TCCCTCTGTAGCCCAGGCTGGCCTcaa................................ | 27 | caa | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGACAGAAGGCCAGGCAGTACCAGTGa.................................................................................................................................................................................................................... | 27 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................TGATACTCTCTGTCAGGACATACCAGG.................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGGTGTTGACAGAAGGCCAGGCAGTACC......................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TCCCTCTGTAGCCCAGGCTGGg..................................... | 22 | g | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................................................................TCCCTCTGTAGCCCAGGCTGGCCTcgc................................ | 27 | cgc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGACAGAAGGCCAGGCAGTACCAGTGT.................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................TGACCTAGTCTCTCATTTTAAGGCTGTA......................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................TGTCAGGACATACCAGGCTGCTCTAGATC...................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................TGTCAGGACATACCAGGCTGCTCTAGAT....................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................AGGCAGTACCAGTGTGGGCGATAAAGG........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................GCTTGAGGGCAAATgatg... | 18 | gatg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TGAATAAGCTTCTCGCTTGAGGGCAAAT....... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................TACTCTCTGTCAGGACATACCAGGCTGC.............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TGGGCGATAAAGGGTCAGTACCTTTGA.......................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................AGACTCCCTCTGTAGCCCAGGCTGGCC.................................... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ...........................................................................................................................................................................................AGACTCCCTCTGTAGCCCAGGCTGGCCTT.................................. | 29 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................AGACTCCCTCTGTAGCCCAGGCTGGCCT................................... | 28 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................CTCCCTCTGTAGCCCAGGCTGGCCTTG................................. | 27 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| ...............................................................................................................................................................................................TCCCTCTGTAGCCCAGGCTGGC..................................... | 22 | 16 | 0.19 | 0.19 | - | - | - | - | 0.12 | - | - | - | - | - | 0.06 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................CCCTCTGTAGCCCAGGCTGGC..................................... | 21 | 18 | 0.06 | 0.06 | - | - | - | - | - | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGAGATGGTGTTGACAGAAGGCCAGGCAGTACCAGTGTGGGCGATAAAGGGTCAGTACCTTTGATACTCTCTGTCAGGACATACCAGGCTGCTCTAGATCAGACAGTAGCCCCAACCTGACCTAGTCTCTCATTTTAAGGCTGTAGGATTATTTCAGCAAACCTTAGCCTTCCTTTTCATGAGACAGAGACTCCCTCTGTAGCCCAGGCTGGCCTTGAATAAGCTTCTCGCTTGAGGGCAAATACGATGA ....................................................................................................(((((((((.......)))..)).))))(((((...(((((((.(((.((........)).)))))))))).......)))))((((((.....)))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) |
|---|