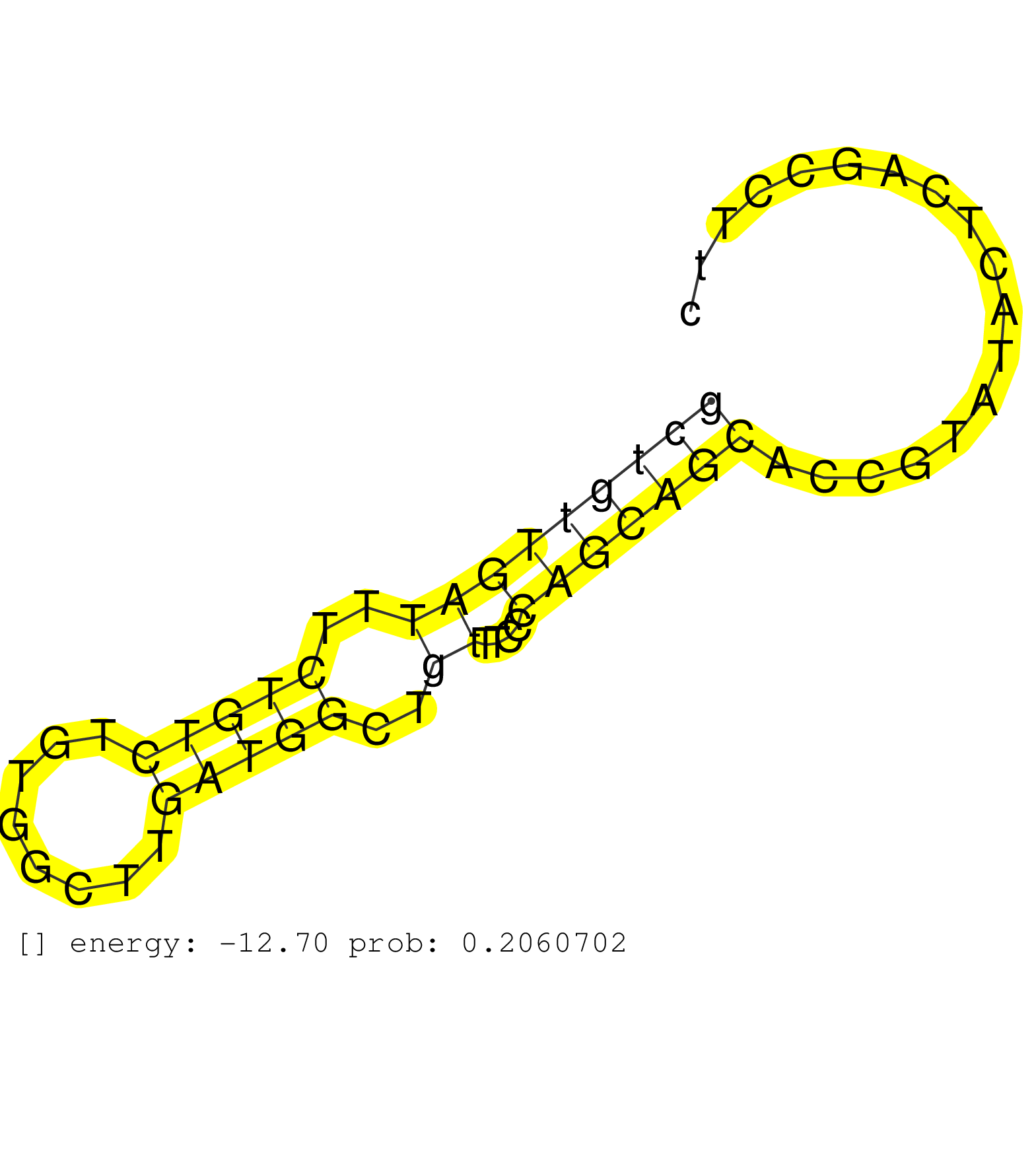

| Gene: 4933437K13Rik | ID: uc007yfj.1_intron_10_0_chr16_11660721_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| CGTCTGTGTTTCTCCGAGGCAAAGCAGCCAATGCGTTCCATGTGTATCAGGTAAGTGCCTGGGCTTTGTGTAGACATCATGGTTGTGGAGCCATGGTGCTTTGCATGTTGGCACCATTTCCTGCCTAGTGCTAGTGTTGCTGTTGATTTCTGTCTGTGGCTTGATGGCTGTTTCCCAGCAGCACCGTATACTCAGCCTTCTGCATGGCTTCAGCCAAGGCCTGGACCATATCATAGTCCTTCTTGGATCT ..........................................................................................................................................(((((((((..(((((........)))))..))....))))))).................................................................... ..........................................................................................................................................139..........................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................TGATTTCTGTCTGTGGCTTGATGGCT................................................................................. | 26 | 1 | 8.00 | 8.00 | 4.00 | 1.00 | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGATTTCTGTCTGTGGCTTGATGGCTGT............................................................................... | 28 | 1 | 6.00 | 6.00 | 1.00 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TTCCCAGCAGCACCGTATACTCAGCCT.................................................... | 27 | 1 | 5.00 | 5.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGATTTCTGTCTGTGGCTTGATGGCTGg............................................................................... | 28 | g | 3.00 | 0.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTGATTTCTGTCTGTGGCTTGATGGC.................................................................................. | 26 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TCCCAGCAGCACCGTATACTCAGCCTT................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TTCCCAGCAGCACCGTATACTCAGCCTT................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....TGTGTTTCTCCGAGGCAAAGCAGCCAAT.......................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TCTGTCTGTGGCTTGATGGCTGTTTt............................................................................ | 26 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................AGCAGCACCGTATACTCAGCCTTCTG................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGTTGATTTCTGTCTGTGGCTTGATGG................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GCCTAGTGCTAGTGTTGCTGTTGATTT..................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................TGCGTTCCATGTGTATCAGGTgcct.................................................................................................................................................................................................. | 25 | gcct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................GGTAAGTGCCTGGGCTTTGTGTAGAC............................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGATTTCTGTCTGTGGCTTGATGGat................................................................................. | 26 | at | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGATTTCTGTCTGTGGCTTGATGGCTa................................................................................ | 27 | a | 1.00 | 8.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................ATTTCTGTCTGTGGCatgc...................................................................................... | 19 | atgc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................AGCCAATGCGTTCCATGTGTATCAGGTA..................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................TTCCATGTGTATCAGGTAAGTGCCTGGGCT......................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................AGCCAATGCGTTCCATGTGTATCAGGa...................................................................................................................................................................................................... | 27 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGATTTCTGTCTGTGGCTTGATGGCTGTa.............................................................................. | 29 | a | 1.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TTCTGCATGGCTTCAtttg.................................. | 19 | tttg | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................GATTTCTGTCTGTGGCTTGATGGCTGTT.............................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGATTTCTGTCTGTGGCTTGATGGCTt................................................................................ | 27 | t | 1.00 | 8.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GCCAATGCGTTCCATGTGTATCAGGTA..................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TAGTGCTAGTGTTGCTGTTGATTTC.................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| CGTCTGTGTTTCTCCGAGGCAAAGCAGCCAATGCGTTCCATGTGTATCAGGTAAGTGCCTGGGCTTTGTGTAGACATCATGGTTGTGGAGCCATGGTGCTTTGCATGTTGGCACCATTTCCTGCCTAGTGCTAGTGTTGCTGTTGATTTCTGTCTGTGGCTTGATGGCTGTTTCCCAGCAGCACCGTATACTCAGCCTTCTGCATGGCTTCAGCCAAGGCCTGGACCATATCATAGTCCTTCTTGGATCT ..........................................................................................................................................(((((((((..(((((........)))))..))....))))))).................................................................... ..........................................................................................................................................139..........................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................CCAGCAGCACCGTATcggt............................................................. | 19 | cggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |