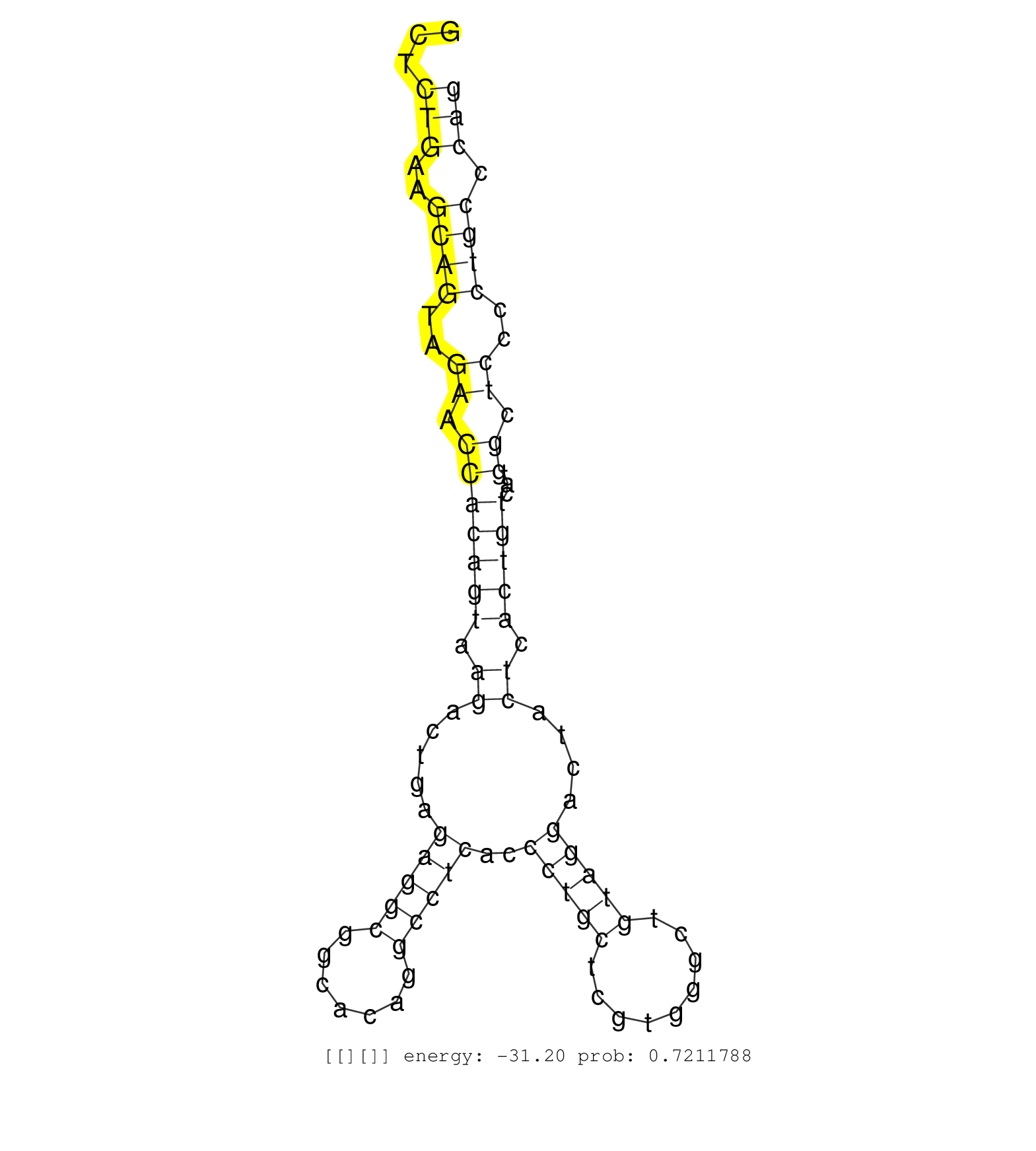

| Gene: Nlrc3 | ID: uc007xzc.1_intron_14_0_chr16_3962541_r.3p | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(1) PIWI.mut |
(8) TESTES |
| AACAGCACAGTTCGCAGGAACCGCCTCAGGTCACCTCAGGTTACATTAGGTTACCCCACAGAGAAGTCTGAGCCCAGTGAAGACGTGAGGCTAGACGTTTGCTCTGAAGCAGTAGAACCACAGTAAGACTGAGAGGCGGCACAGGCCTCACCCTGCTCGTGGGCTGTAGGACTACTCACTGTCATGGCTCCCCTGCCCAGGCTGGACAACAACCAGTTCCAGGACCCTGTGATGGAGTTGCTGGGCAGCG .......................................................................................................(((..((((..((.(((((((.((.....(((((.......)))))..(((((.........)))))....)).)))))...)).))..)))).))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................TAGACGTTTGCTCTGAAGCAGTAGAACC................................................................................................................................... | 28 | 1 | 8.00 | 8.00 | 1.00 | 4.00 | 3.00 | - | - | - | - | - |
| ......................................................................................................TCTGAAGCAGTAGAACCACAGTAAGACT........................................................................................................................ | 28 | 1 | 5.00 | 5.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | - |
| ......................................................................................GAGGCTAGACGTTTGCTCTGAAGCAG.......................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - |
| ...........................................................................................TAGACGTTTGCTCTGAAGCAGTAGAAC.................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................GCAGTAGAACCACAGTAAGACTGAGAGG.................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................TGAAGCAGTAGAACCACAGTAAGACTG....................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................GACGTTTGCTCTGAAGCAGTAGAACCA.................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................ACAGTAAGACTGAGAGGCGGCACAGGCC....................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................GAAGCAGTAGAACCACAGTAAGACTGAG..................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................TGAAGCAGTAGAACCACAGTAAGACT........................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................TGCTCTGAAGCAGTAGAACCACAGTAAGA.......................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................GGCTAGACGTTTGCTCTGAAGCAGTA........................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................TGCTCTGAAGCAGTAGAACCACAGTAAG........................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| AACAGCACAGTTCGCAGGAACCGCCTCAGGTCACCTCAGGTTACATTAGGTTACCCCACAGAGAAGTCTGAGCCCAGTGAAGACGTGAGGCTAGACGTTTGCTCTGAAGCAGTAGAACCACAGTAAGACTGAGAGGCGGCACAGGCCTCACCCTGCTCGTGGGCTGTAGGACTACTCACTGTCATGGCTCCCCTGCCCAGGCTGGACAACAACCAGTTCCAGGACCCTGTGATGGAGTTGCTGGGCAGCG .......................................................................................................(((..((((..((.(((((((.((.....(((((.......)))))..(((((.........)))))....)).)))))...)).))..)))).))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|