
| Gene: Itga5 | ID: uc007xyb.1_intron_7_0_chr15_103180886_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.mut |
(11) TESTES |
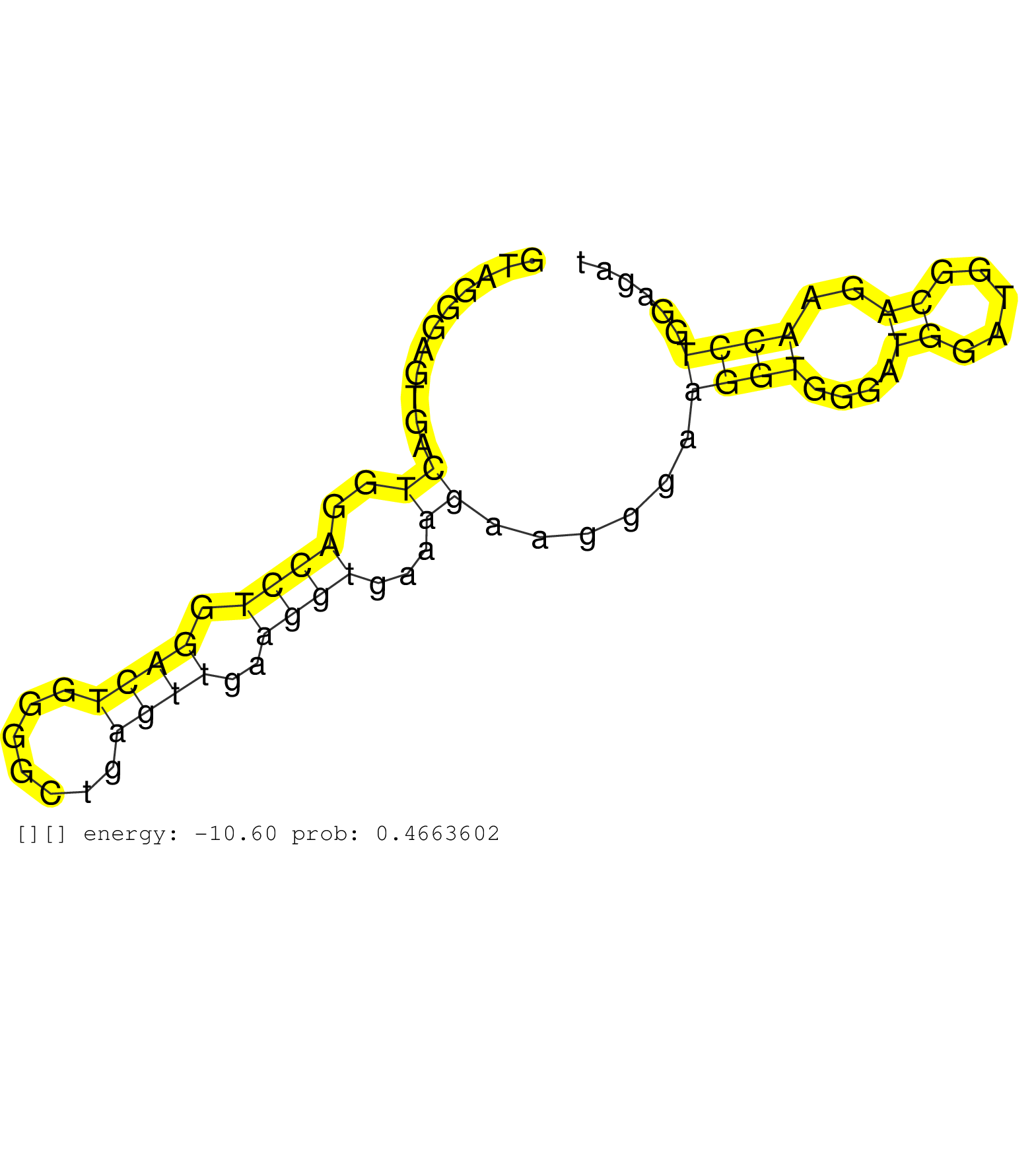
| CATCTTCAAGACACAAAGAAAACCATCCAGTTTGACTTTCAGATCCTCAGGTAGGGAGTGACTGGACCTGGACTGGGGCTGAGTTGAAGGTGAAAGAAGGGAAGGTGGGATGGATGGCAGAACCTGGAGATTCTTGTCAAACTGCTTGCCTCTTGCCTCCATCCCACAGCAAGAACCTGAACAACTCACAAAGCAACGTGGTCTCCTTCCCACTCTCGG .............................................................((..((((.((((.......))))..))))...))......((((....((.....))..)))).............................................................................................. ..................................................51..............................................................................131...................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT2() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................GGTGGGATGGATGGCAGAACCTGG............................................................................................ | 24 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTAGGGAGTGACTGGACCTGGACTGGGGC............................................................................................................................................ | 29 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ....TTCAAGACACAAAGAAga..................................................................................................................................................................................................... | 18 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTAGGGAGTGACTGGACCTGGACTGt............................................................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................AGGTGGGATGGATGGCAGAACCTGG............................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................GTGGGATGGATGGCAGAACC............................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................TGGGATGGATGGCAGAACCTGG............................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................TGGGATGGATGGCAGAACCTGGA........................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................GGTGGGATGGATGGCAGAACCTG............................................................................................. | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGGGATGGATGGCAGAACCTGGAGA......................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................AGGGAGTGACTGGACCTGGACTGGGGC............................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................CTGGACTGGGGCTGAG........................................................................................................................................ | 16 | 7 | 0.14 | 0.14 | - | - | - | - | 0.14 | - | - | - | - | - | - |
| CATCTTCAAGACACAAAGAAAACCATCCAGTTTGACTTTCAGATCCTCAGGTAGGGAGTGACTGGACCTGGACTGGGGCTGAGTTGAAGGTGAAAGAAGGGAAGGTGGGATGGATGGCAGAACCTGGAGATTCTTGTCAAACTGCTTGCCTCTTGCCTCCATCCCACAGCAAGAACCTGAACAACTCACAAAGCAACGTGGTCTCCTTCCCACTCTCGG .............................................................((..((((.((((.......))))..))))...))......((((....((.....))..)))).............................................................................................. ..................................................51..............................................................................131...................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT2() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................CACAGCAAGAACCTGAACAACTCACA............................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GGAGTGACTGGACtgcc........................................................................................................................................................ | 17 | tgcc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................TTCAGATCCTCAGGT....................................................................................................................................................................... | 15 | 19 | 0.05 | 0.05 | - | - | - | - | - | - | - | - | - | - | 0.05 |