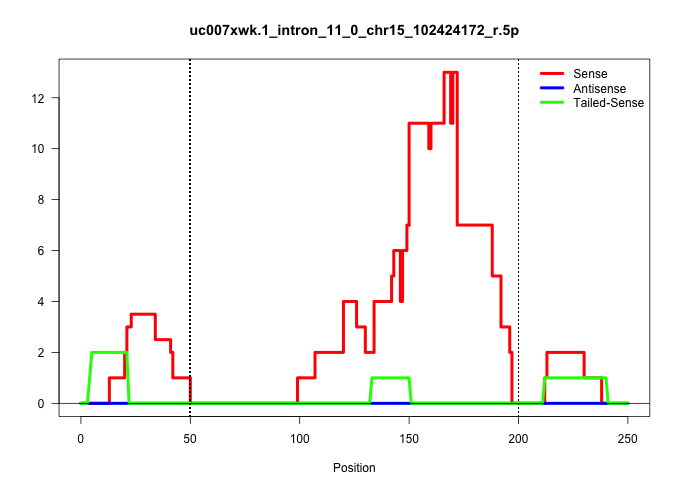
| Gene: Atf7 | ID: uc007xwk.1_intron_11_0_chr15_102424172_r.5p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(2) OVARY |
(3) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| CTTTTGGAGGAAGGCGGGGCCAGAGGACTGAAGTGTGGGGAGAACTTGGGGTAAGTAGCCCCGGGGGGCACAGGGCGTGGCGAGGAAAGGGGGGTTGAATCCTGAGGGAAGCGAAGTCTAGTGGAAGTCTAATGGAGGGATCTTGGAGTGGAAGATAGAGGAGGACTGCTAAACTAGGTTGAAGACTCAAAAGATTTCCCAAAAAGTGGGTTTAGGGCTTGGCACTGCAGAAGGCAGAGGTAAATAGGCA |
Size | Perfect hit | Total Norm | Perfect Norm | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR037899(GSM510435) ovary_rep4. (ovary) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................GAAGATAGAGGAGGACTGCTAA.............................................................................. | 22 | 1 | 4.00 | 4.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................................TGCTAAACTAGGTTGAAGACTCAAAA.......................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - |
| ..........................................................................................................................................................................AAACTAGGTTGAAGACTCAAAAGATTT..................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................GAGGGATCTTGGAGTGGAAGATAGA........................................................................................... | 25 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................GTGGAAGTCTAATGGAGGGATCTTGG........................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .....GGAGGAAGGCGGGGaca.................................................................................................................................................................................................................................... | 17 | aca | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................GGAAGATAGAGGAGGACTGCTAA.............................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................GAAGCGAAGTCTAGTGGAAGTCT........................................................................................................................ | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GTGGAAGATAGAGGAGGACTGC................................................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TCCTGAGGGAAGCGAAGTCTAGTGGAA............................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................AGAGGACTGAAGTGTGGGGAG................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................AGGGCTTGGCACTGCAG.................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....TGGAGGAAGGCGGGGacg.................................................................................................................................................................................................................................... | 18 | acg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GGAGGACTGCTAAACTAGGTTGAAGACTC.............................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TGGAGTGGAAGATAGAGGAGGACTGC................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................CAGAGGACTGAAGTGTGGGGAGAACTTGGG........................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................GAGGACTGCTAAACTAGGTTGAAGACTC.............................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TAAACTAGGTTGAAGACTCAAAAGATT...................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................GGAGGGATCTTGGAGaaa................................................................................................... | 18 | aaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................................................................................TAGGGCTTGGCACTGCAGAAGGCAGA............ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............GCGGGGCCAGAGGACTGAAGT........................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GTGGAAGATAGAGGAGGACTGCTAA.............................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................................................................................................................................................................................TAGGGCTTGGCACTGCAGAAGGCAGAatt......... | 29 | att | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTGGAGTGGAAGATAGAGGAGGACTGC................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................AGGACTGAAGTGTGGGGA................................................................................................................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| CTTTTGGAGGAAGGCGGGGCCAGAGGACTGAAGTGTGGGGAGAACTTGGGGTAAGTAGCCCCGGGGGGCACAGGGCGTGGCGAGGAAAGGGGGGTTGAATCCTGAGGGAAGCGAAGTCTAGTGGAAGTCTAATGGAGGGATCTTGGAGTGGAAGATAGAGGAGGACTGCTAAACTAGGTTGAAGACTCAAAAGATTTCCCAAAAAGTGGGTTTAGGGCTTGGCACTGCAGAAGGCAGAGGTAAATAGGCA |
Size | Perfect hit | Total Norm | Perfect Norm | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR037899(GSM510435) ovary_rep4. (ovary) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|