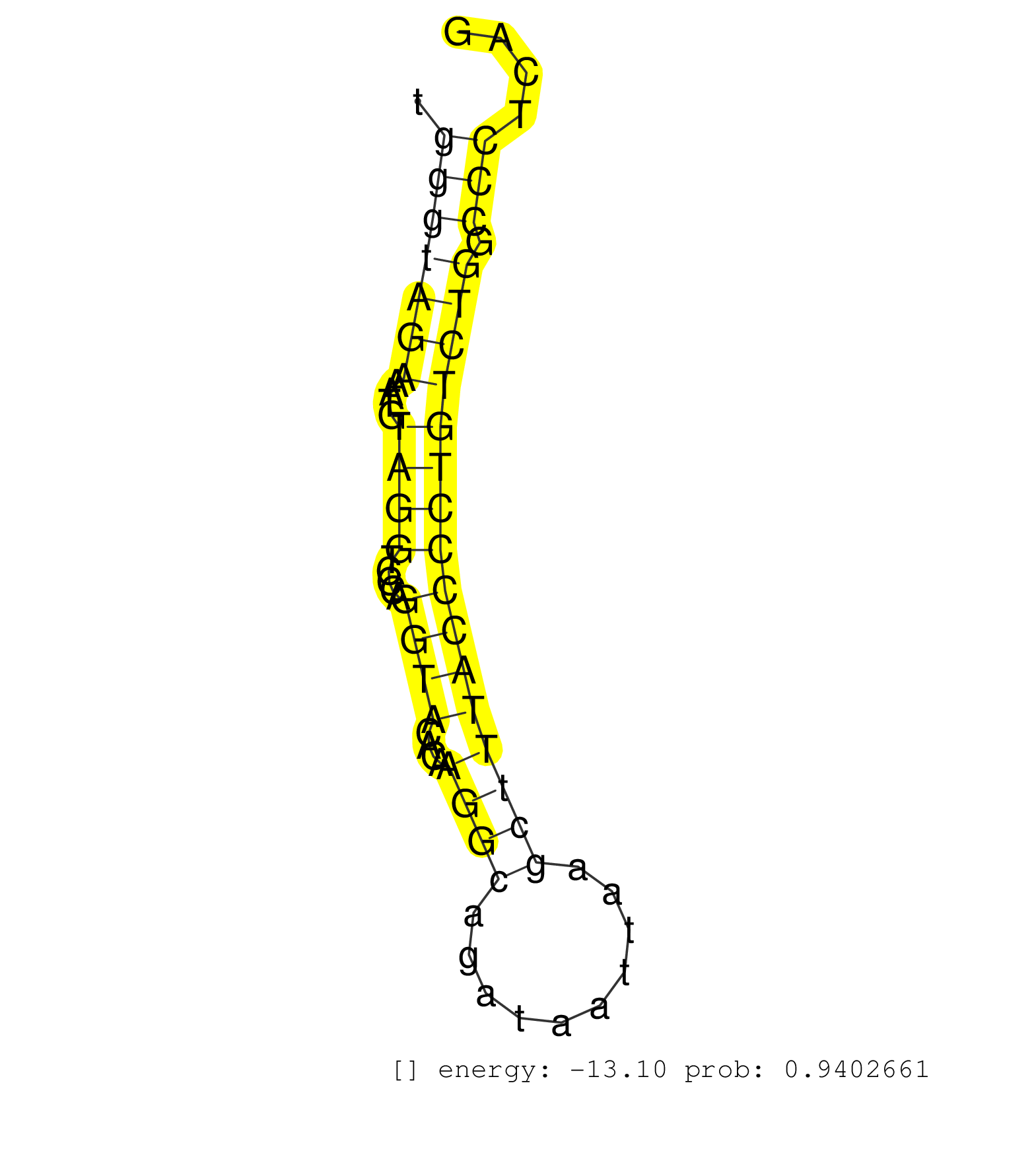

| Gene: Map3k12 | ID: uc007xwc.1_intron_10_0_chr15_102334738_r | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(3) PIWI.ip |
(12) TESTES |
| GACATCAAGCATCTTCGAAAGCTGAAGCACCCCAACATCATCACTTTCAAGTGAGGTCCAGGGCAGGGGCTGGGGGTACCATGACACATAGGACTCCCCTCACCTGGGACTATATCCCCTTTCTCAAAGGAGACCCAGAAATGGCATGGGTAGAAATGTAGGTCCCAGGTACACAAGGCAGATAATTAAGCTTTACCCCTGTCTGGCCCTCAGGGGTGTTTGCACACAGGCCCCCTGCTACTGCATCCTTATGGAATTCTGCG ...................................................................................................................................................(((((((....((((.....((((....((((..........)))))))))))))))).)))...................................................... ..................................................................................................................................................147...............................................................213................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................TTACCCCTGTCTGGCCCTCAGt................................................. | 22 | t | 14.00 | 0.00 | 3.00 | 5.00 | 1.00 | - | 2.00 | - | 2.00 | - | - | 1.00 | - | - |
| ................................................................................................................................................................................................TTACCCCTGTCTGGCCCTCAGa................................................. | 22 | a | 4.00 | 0.00 | - | - | 1.00 | - | - | 3.00 | - | - | - | - | - | - |
| ...............CGAAAGCTGAAGCACCCCAACATCATC............................................................................................................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TTACCCCTGTCTGGCCCTCAGag................................................ | 23 | ag | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................AGAAATGTAGGTCCCAGGTACACAAGG..................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................AGAAATGTAGGTCCCAGGTACACAAGGCA................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - |
| ...............................................................................................................................................................................................TTTACCCCTGTCTGGCCCTCAGt................................................. | 23 | t | 2.00 | 0.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TTACCCCTGTCTGGCCCTCAGta................................................ | 23 | ta | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TTACCCCTGTCTGGCCCTCAGttat.............................................. | 25 | ttat | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GAAATGGCATGGGTAGAAATGTAGGTC................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............TCGAAAGCTGAAGCACCCCAACATCAT.............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................................................................................................................................TTACCCCTGTCTGGCCCTCAGattt.............................................. | 25 | attt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................................................................................................................TTACCCCTGTCTGGCCCTCAGtt................................................ | 23 | tt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................AGAAATGTAGGTCCCAGGTACACAAGGC.................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................................CCCTGTCTGGCCCTCAGt................................................. | 18 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TTACCCCTGTCTGGCCCTCAaa................................................. | 22 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............CGAAAGCTGAAGCACCCCAACATCAT.............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................................................................TTACCCCTGTCTGGCCCa..................................................... | 18 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................GATAATTAAGCTTTACCCCTGTCTGGCa....................................................... | 28 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................TGACACATAGGACTCCCCTCACCTGGGACTAT...................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| GACATCAAGCATCTTCGAAAGCTGAAGCACCCCAACATCATCACTTTCAAGTGAGGTCCAGGGCAGGGGCTGGGGGTACCATGACACATAGGACTCCCCTCACCTGGGACTATATCCCCTTTCTCAAAGGAGACCCAGAAATGGCATGGGTAGAAATGTAGGTCCCAGGTACACAAGGCAGATAATTAAGCTTTACCCCTGTCTGGCCCTCAGGGGTGTTTGCACACAGGCCCCCTGCTACTGCATCCTTATGGAATTCTGCG ...................................................................................................................................................(((((((....((((.....((((....((((..........)))))))))))))))).)))...................................................... ..................................................................................................................................................147...............................................................213................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......AGCATCTTCGAAAGCTGAAGCACCCCAACAT................................................................................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TCCCAGGTACACAAgc....................................................................................... | 16 | gc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |