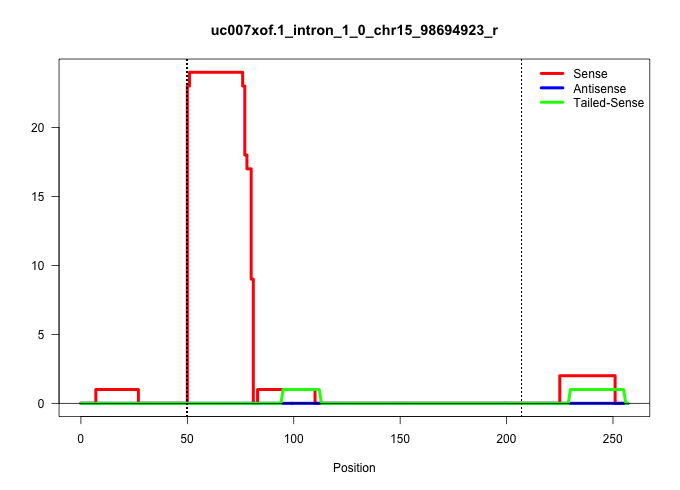
| Gene: BC012717 | ID: uc007xof.1_intron_1_0_chr15_98694923_r | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(12) TESTES |
| AGCCAGCCTGTCACGTCTGTTGCCGAGCAGCACCCAGCTGTCTGTAGCAGGTAAGTGGTGTCAACTGAGATTGGGGCGGGGCATTGGGAGCGGTGAGCCTGGAGGGGCCATGTTAAACAGGCAGCCAGTCAGCAGTGCAGTCCACCTCGTTGATGGAGTTGTTAGCTGGAGAGAGCCACAGTGCCTGCTCCCCTGTGCCCTCTGCAGATTGTCCCCCCCAGAGCCTGGTGAGATCCCCATTGATGCGCCGGATGCTC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTGGTGTCAACTGAGATTGGGGCGGGG................................................................................................................................................................................ | 31 | 1 | 9.00 | 9.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGGTGTCAACTGAGATTGGGGCGGG................................................................................................................................................................................. | 30 | 1 | 8.00 | 8.00 | 7.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTAAGTGGTGTCAACTGAGATTGGGGC.................................................................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | - | 1.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TGGTGAGATCCCCATTGATGCGCCGG...... | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .......CTGTCACGTCTGTTGCCGAG...................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................AGCCTGGAGGGGCCcgga................................................................................................................................................ | 18 | cgga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTAAGTGGTGTCAACTGAGATTGGGGCG................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................TTGGGAGCGGTGAGCCTGGAGGGGCCA................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................TAAGTGGTGTCAACTGAGATTGGGG..................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................AGATCCCCATTGATGCGCCGGATGgt. | 26 | gt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| AGCCAGCCTGTCACGTCTGTTGCCGAGCAGCACCCAGCTGTCTGTAGCAGGTAAGTGGTGTCAACTGAGATTGGGGCGGGGCATTGGGAGCGGTGAGCCTGGAGGGGCCATGTTAAACAGGCAGCCAGTCAGCAGTGCAGTCCACCTCGTTGATGGAGTTGTTAGCTGGAGAGAGCCACAGTGCCTGCTCCCCTGTGCCCTCTGCAGATTGTCCCCCCCAGAGCCTGGTGAGATCCCCATTGATGCGCCGGATGCTC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|