
| Gene: Fkbp11 | ID: uc007xnm.1_intron_3_0_chr15_98557730_r | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(3) PIWI.mut |
(20) TESTES |
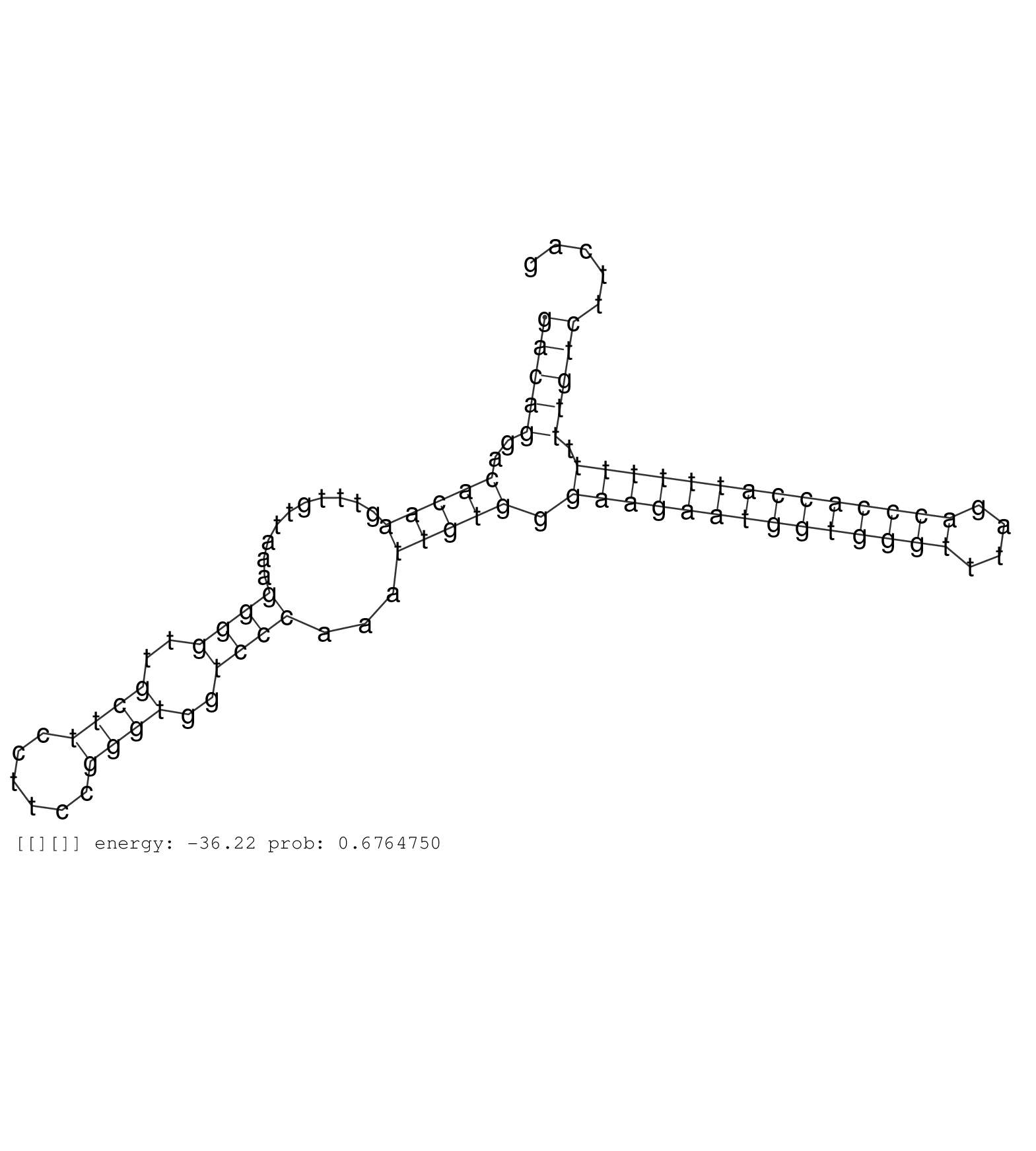
| CGTGCACGGAATCTGCTGCCATTGGAGACACGCTCCACATACACTACACGGTGAGGGGCTTGGGGGTCCGGGGCGTGGCCTGGGAGGGGCGGCGCCCGGCTGAAACACGTCAGCTTTGGCTTGGACACGTGGCAAGCTATCCTAGAGAACTTGAGTCCTAGGATCAGCCCGCACGTGGGGGCGTCCTCTCGCCTGCTTCTCTTTCTTCCTTGTTTTCTGCCGATTCCTCTCCAGCGCTGAGAAAAGAAGGATAGAGAGAGCTCCAAGAAGCGCCAGCCCCGTGGGGCTTCCTGTTCCTTCTTGCCCATGGTGGTGGTGGGTGGAAGGAGATGGGGACAGGACACAAGTTTGTTAAAGGGGTTGCTTCCTTCCGGGTGGTCCCAAATTGTGGGAAGAATGGTGGGTTTAGACCCACCATTTTTTTTTGTCTTCAGGGCAGTTTGGTAGATGGACGCATTATTGACACTTCTCTGACCAGAGATCC ..............................................................................................................................................................................................................................................................................................................................................(((((..(((((..........((((..((((......))))..))))...))))).((((((((((((((....)))))))))))))).)))))....................................................... ..............................................................................................................................................................................................................................................................................................................................................335................................................................................................434................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................TGGTAGATGGACGCATTATTGACACTT................ | 27 | 1 | 13.00 | 13.00 | 4.00 | 4.00 | 2.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................TGGTAGATGGACGCATTATTGACACT................. | 26 | 1 | 13.00 | 13.00 | 2.00 | 3.00 | 2.00 | 2.00 | 1.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................TGGTAGATGGACGCATTATTGACACTTC............... | 28 | 1 | 4.00 | 4.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................TCCAGCGCTGAGAAAAGAAGGATAGA..................................................................................................................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................TAGAGAGAGCTCCAAGAAGCGCCAGCCCC............................................................................................................................................................................................................ | 29 | 1 | 3.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................TCCAGCGCTGAGAAAAGAAGGATAG...................................................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................GGATAGAGAGAGCTCCAAGAAGCGCCA................................................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................AGATGGACGCATTATTGACACTTCTCTG........... | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................................................GATGGACGCATTATTGACACTTCTCTG........... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................AGGATAGAGAGAGCTCCAAGAAGCGCCA................................................................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................AAGGATAGAGAGAGCTCCAAGAAGCGCC.................................................................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................TGAGAAAAGAAGGATAGAGAGAGCT.............................................................................................................................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................TGAGAAAAGAAGGATAGAGAGAGCTCC............................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................TAGATGGACGCATTATTGACACTTCTC............. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................GACGCATTATTGACACTT................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................GGTAGATGGACGCATTATTGACACTTCTC............. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TGCACGGAATCTGCTGCCATTGGAGACA...................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................GGATAGAGAGAGCTCCAAGAAGCGCC.................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................AGGATAGAGAGAGCTCCAAGAAGCGCC.................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................................................................................................................................AGGATAGAGAGAGCTCCAAGAAGCGCCAGC............................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................CAGGGCAGTTTGGTAGATGGACGCA............................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................................................................................................................................................AGGATAGAGAGAGCTCCAAGAAGCGC................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................GTTTGGTAGATGGACGCATTATTGAC.................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACGCATTATTGACACTTCTCTGACCAGA..... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................TAGAGAGAGCTCCAAGAAGCGCCAGC............................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................TTGGTAGATGGACGCATTATTGACACT................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGGACGCATTATTGACACTTCTCTGA.......... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................................................................................TCTCCAGCGCTGAGAAAAGAAGGATAG...................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................ATAGAGAGAGCTCCAAGA........................................................................................................................................................................................................................ | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| CGTGCACGGAATCTGCTGCCATTGGAGACACGCTCCACATACACTACACGGTGAGGGGCTTGGGGGTCCGGGGCGTGGCCTGGGAGGGGCGGCGCCCGGCTGAAACACGTCAGCTTTGGCTTGGACACGTGGCAAGCTATCCTAGAGAACTTGAGTCCTAGGATCAGCCCGCACGTGGGGGCGTCCTCTCGCCTGCTTCTCTTTCTTCCTTGTTTTCTGCCGATTCCTCTCCAGCGCTGAGAAAAGAAGGATAGAGAGAGCTCCAAGAAGCGCCAGCCCCGTGGGGCTTCCTGTTCCTTCTTGCCCATGGTGGTGGTGGGTGGAAGGAGATGGGGACAGGACACAAGTTTGTTAAAGGGGTTGCTTCCTTCCGGGTGGTCCCAAATTGTGGGAAGAATGGTGGGTTTAGACCCACCATTTTTTTTTGTCTTCAGGGCAGTTTGGTAGATGGACGCATTATTGACACTTCTCTGACCAGAGATCC ..............................................................................................................................................................................................................................................................................................................................................(((((..(((((..........((((..((((......))))..))))...))))).((((((((((((((....)))))))))))))).)))))....................................................... ..............................................................................................................................................................................................................................................................................................................................................335................................................................................................434................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................TTGTCTTCAGGGCAgata.............................................. | 18 | gata | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................................................................................................................................................................TCTGCCGATTCCTCTCCAGCGCTGAGA.................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |