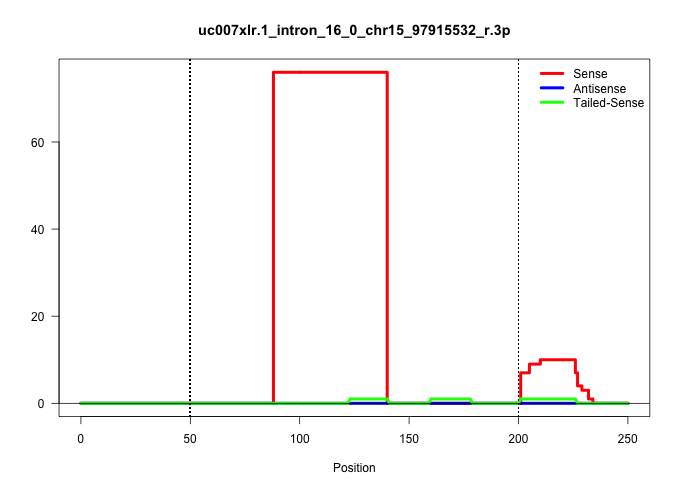
| Gene: Senp1 | ID: uc007xlr.1_intron_16_0_chr15_97915532_r.3p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(11) TESTES |
| ATTCTTGTGTAGGTTTTGAGCTGCTTGTTAGATGCTCTCAGTCAATTCTAAATGGAGCAGTGTCTTCACATGCTAGGAAGGAGTGGTATTATTTGTCTGTATAATTCCTCTCTGAGAACAGTGTTTTAGGGGAAAAGATTGCCTCACAGGAAAGTGTTGTTGAAGGGCTGTGTCTTGCTCTAATCTGACTCTTCCCCTAGATGACACAGCTGATGGGGTGAAGATGGACGCTGGGGAGGTGACCTTAGTG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR037903(GSM510439) testes_rep4. (testes) | mjTestesWT1() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................TTATTTGTCTGTATAATTCCTCTCTGAGAACAGTGTTTTAGGGGAAAAGATT.............................................................................................................. | 52 | 1 | 76.00 | 76.00 | 76.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGACACAGCTGATGGGGTGAAGATGG....................... | 26 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - |
| .........................................................................................................................................................................................................TGACACAGCTGATGGGGTGAAGATG........................ | 25 | 1 | 3.00 | 3.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................................................................................ACAGCTGATGGGGTGAAGATGGACGCT.................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................................................TGATGGGGTGAAGATGGACGCTGG................ | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TTTTAGGGGAAAAGtctt............................................................................................................. | 18 | tctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................................................................................................TGACACAGCTGATGGGGTGAAGATGa....................... | 26 | a | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGACACAGCTGATGGGGTGAAGATGGAC..................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGAAGGGCTGTGTCTagat....................................................................... | 19 | agat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ATTCTTGTGTAGGTTTTGAGCTGCTTGTTAGATGCTCTCAGTCAATTCTAAATGGAGCAGTGTCTTCACATGCTAGGAAGGAGTGGTATTATTTGTCTGTATAATTCCTCTCTGAGAACAGTGTTTTAGGGGAAAAGATTGCCTCACAGGAAAGTGTTGTTGAAGGGCTGTGTCTTGCTCTAATCTGACTCTTCCCCTAGATGACACAGCTGATGGGGTGAAGATGGACGCTGGGGAGGTGACCTTAGTG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR037903(GSM510439) testes_rep4. (testes) | mjTestesWT1() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|