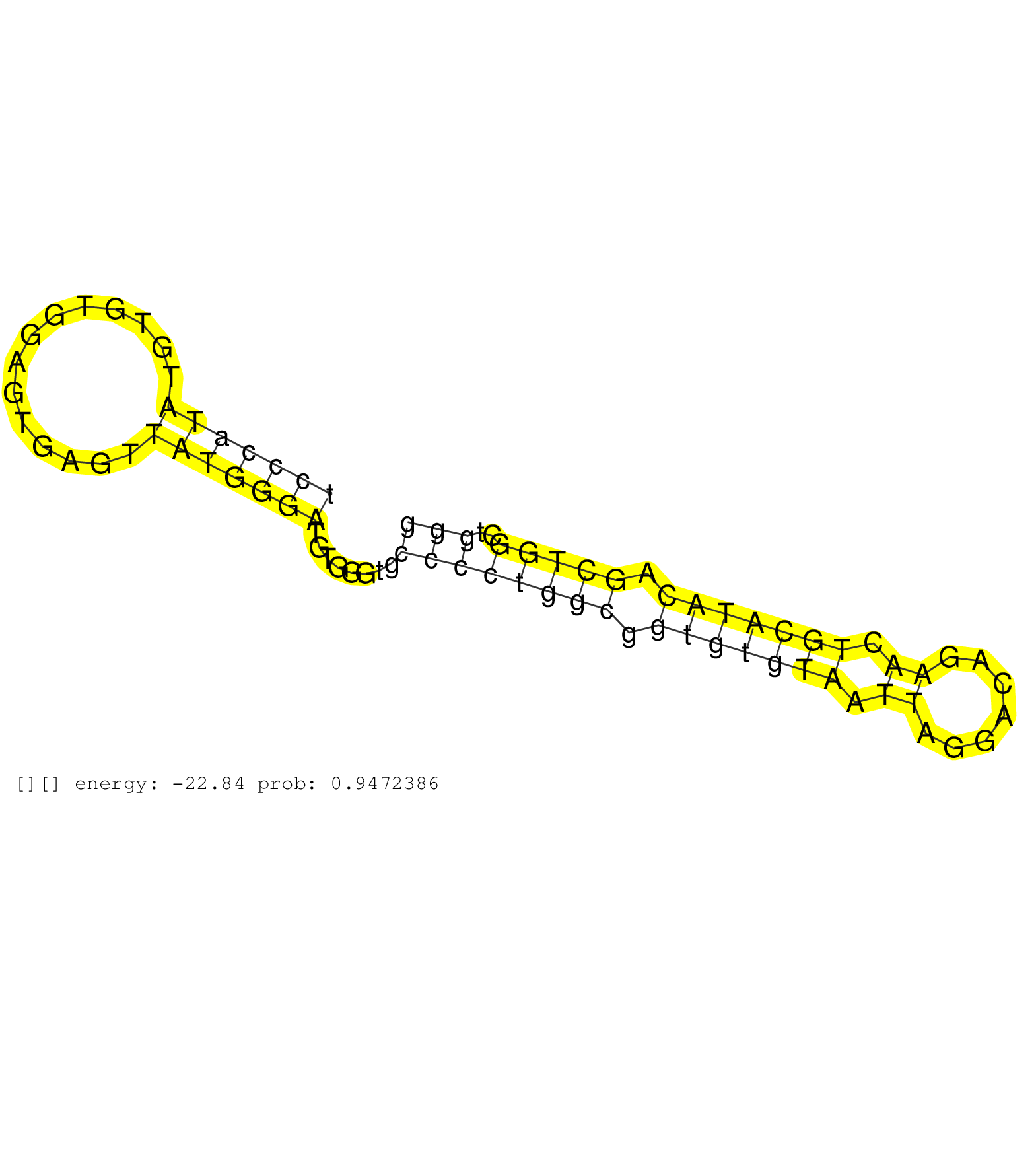
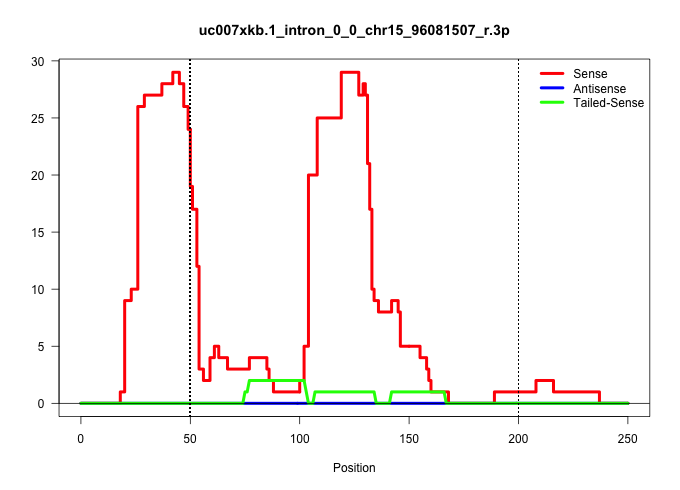
| Gene: AK014752 | ID: uc007xkb.1_intron_0_0_chr15_96081507_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(22) TESTES |
| GGAGGCATGATTTGCCCAGTTAACTCTGGAGAAGCATTGAGCTGAAGAACCGGTTCCCATATGTGTGGAGTGAGTTATGGGATGTGGGTGCCCCTGGCGGTGTGTAATTAGGACAGAACTGCATACAGCTGGCTGGGAAGAATGGCTGGATCGTCAGGTGATCGTCCTACCACCCTCCCTGTGCCCTCCTTCTATCTAAGGAGCAGGCTCTTCGGTGGAGACCTTTTGGGAGGAGGCCCAGCCGACCGGA ......................................................(((((((..............)))))))........((((((((.(((((((.((.......)).))))))).)))))..)))................................................................................................................. ......................................................55................................................................................137............................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................TACAGCTGGCTGGGAAGAATGGCa....................................................................................................... | 24 | a | 10.00 | 0.00 | - | - | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TGGAGAAGCATTGAGCTGAAGAACCGGT.................................................................................................................................................................................................... | 28 | 1 | 9.00 | 9.00 | - | 1.00 | - | 1.00 | 1.00 | 2.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................TAATTAGGACAGAACTGCATACAGCTGGC..................................................................................................................... | 29 | 1 | 7.00 | 7.00 | 1.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................TGGAGAAGCATTGAGCTGAAGAACCGG..................................................................................................................................................................................................... | 27 | 1 | 7.00 | 7.00 | - | 1.00 | - | - | 2.00 | - | 1.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TAATTAGGACAGAACTGCATACAGCTGG...................................................................................................................... | 28 | 1 | 5.00 | 5.00 | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................TAATTAGGACAGAACTGCATACAGCTG....................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................TAACTCTGGAGAAGCATTGAGCTGAAGAAC........................................................................................................................................................................................................ | 30 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................TAGGACAGAACTGCATACAGCTGGC..................................................................................................................... | 25 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGCATACAGCTGGCTGGGAAGAATGGC........................................................................................................ | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................TGGAGAAGCATTGAGCTGAAGAACC....................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................TAACTCTGGAGAAGCATTGAGCTGAAG........................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TAACTCTGGAGAAGCATTGAGCTGAAGAA......................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGTAATTAGGACAGAACTGCATACAGCTG....................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATGTGTGGAGTGAGTTATGGGATGTGGG.................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................................................................TCTTCGGTGGAGACCTTTTGGGAGGAGGC............. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGGGATGTGGGTGCCCCTGGCGGTGg................................................................................................................................................... | 26 | g | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................CTCTGGAGAAGCATTGAGCTGAAGAAC........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TAGGACAGAACTGCATACAGCTGGCT.................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGGCTGGATCGTCAGGTGATCGTCCT.................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGGGAAGAATGGCTGGATCGTCAGGTG.......................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGTAATTAGGACAGAACTGCATACA........................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGGGAAGAATGGCTGGATCGTCAGGT........................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TAGGACAGAACTGCAgaa............................................................................................................................ | 18 | gaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................AGAAGCATTGAGCTGAAGAACCGGT.................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TATCTAAGGAGCAGGCTCTTCGGTGG................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................TGGAGAAGCATTGAGCTGAAGAACCGGTTC.................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................TATGGGATGTGGGTGCCCCTGGCGGggtg.................................................................................................................................................. | 29 | ggtg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGCATACAGCTGGCTGGGAAGAATGG......................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................TTAGGACAGAACTGCATACAGCTGGCTt................................................................................................................... | 28 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TTCTATCTAAGGAGCAGGCTCTTCGGT.................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TGAGCTGAAGAACCGGTTCCCATATG........................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGGGATGTGGGTGCCCCTGGCGGTG.................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TAGGACAGAACTGCATACAGCTGGCTGG.................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TGAAGAACCGGTTCCCATATGTGTG....................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TTAGGACAGAACTGCATACAGCTGGC..................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGTAATTAGGACAGAACTGCATACAG.......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................GTTAACTCTGGAGAAGCATTGAGCTGA............................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGTAATTAGGACAGAACTGCATACAGCT........................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................TGTGTAATTAGGACAGAACTGCATACA........................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGGGAAGAATGGCTGGATCGTCAGG............................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TAGGACAGAACTGCATACAGCTGGCTG................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TGGCTGGGAAGAATGGCTGGATCGTC............................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGGCTGGATCGTCAGGTGATCGTCa................................................................................... | 25 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TGTGTGGAGTGAGTTATGGGATGTG.................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TATGTGTGGAGTGAGTTATGGGATGT..................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGAGGCATGATTTGCCCAGTTAACTCTGGAGAAGCATTGAGCTGAAGAACCGGTTCCCATATGTGTGGAGTGAGTTATGGGATGTGGGTGCCCCTGGCGGTGTGTAATTAGGACAGAACTGCATACAGCTGGCTGGGAAGAATGGCTGGATCGTCAGGTGATCGTCCTACCACCCTCCCTGTGCCCTCCTTCTATCTAAGGAGCAGGCTCTTCGGTGGAGACCTTTTGGGAGGAGGCCCAGCCGACCGGA ......................................................(((((((..............)))))))........((((((((.(((((((.((.......)).))))))).)))))..)))................................................................................................................. ......................................................55................................................................................137............................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|