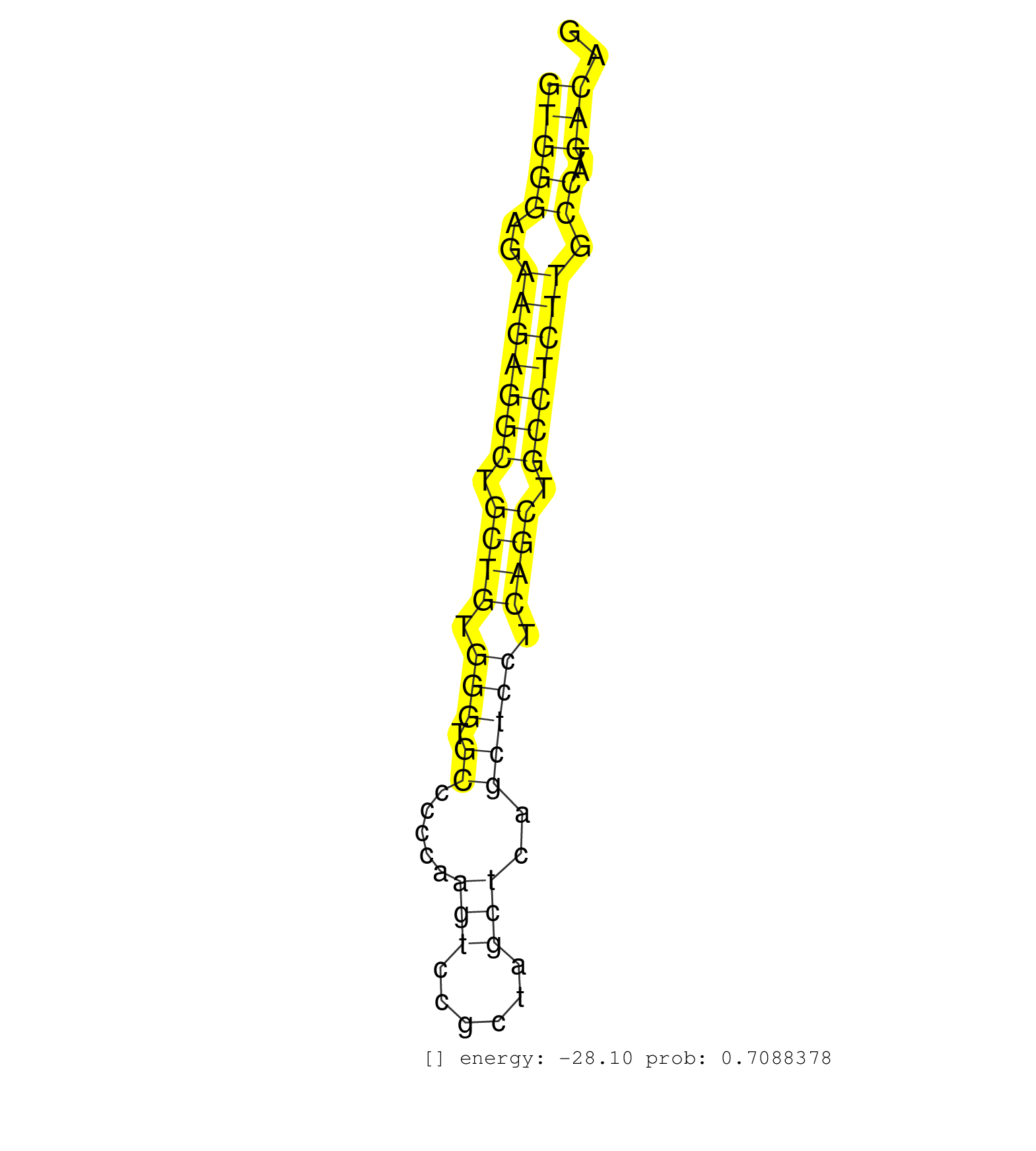

| Gene: 1700027J05Rik | ID: uc007xfs.1_intron_2_0_chr15_89015895_r | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
| GGACCCCAGCTCACCTGCATCCTCAAGGGTGACTGGCTGGGCCTCTACAGGTGGGAGAAGAGGCTGCTGTGGGTGCCCCCAAGTCCGCTAGCTCAGCTCCTCAGCTGCCTCTTGCCATCACAGGCGGTTTTTCAAGTCCCCCCATTTTGATGGCTGGTATCGGCAACGACACA ..................................................(((((..(((((((.((((.(((.((.....(((......)))..))))).)))).))))))).))..))).................................................... ..................................................51......................................................................123................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................TCAGCTGCCTCTTGCCATCACAGt................................................. | 24 | t | 11.00 | 9.00 | 7.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - |
| ....................................................................................................TCAGCTGCCTCTTGCCATCACAG.................................................. | 23 | 1 | 9.00 | 9.00 | 8.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TCAGCTGCCTCTTGCCATCA..................................................... | 20 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................TCAGCTGCCTCTTGCCATCACA................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TCAGCTGCCTCTTGCCATCAtagt................................................. | 24 | tagt | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGAGAAGAGGCTGCTGTGGGTGC................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................TGGGAGAAGAGGCTGCTGTGGGTGC................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................CTCAAGGGTGACTGcgta...................................................................................................................................... | 18 | cgta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTGGGAGAAGAGGCTGCTGTG...................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTGGGAGAAGAGGCTGCTGTGGGTG.................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................CAAGGGTGACTGGCTGGGCCTCTACA............................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGATGGCTGGTATCGGCAACGACACAatt | 29 | att | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| GGACCCCAGCTCACCTGCATCCTCAAGGGTGACTGGCTGGGCCTCTACAGGTGGGAGAAGAGGCTGCTGTGGGTGCCCCCAAGTCCGCTAGCTCAGCTCCTCAGCTGCCTCTTGCCATCACAGGCGGTTTTTCAAGTCCCCCCATTTTGATGGCTGGTATCGGCAACGACACA ..................................................(((((..(((((((.((((.(((.((.....(((......)))..))))).)))).))))))).))..))).................................................... ..................................................51......................................................................123................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|