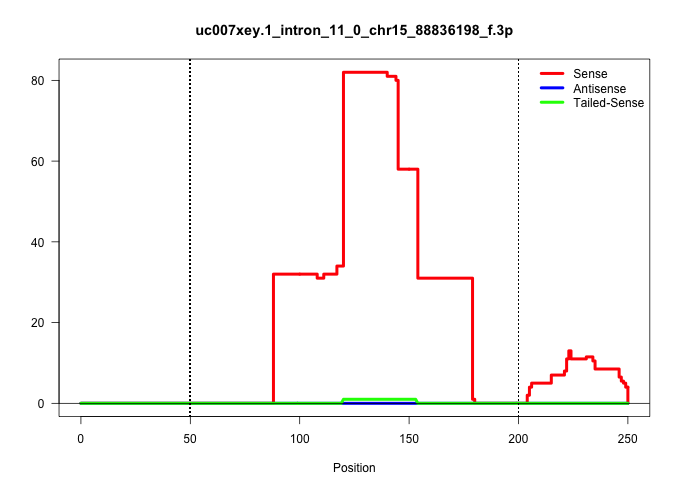
| Gene: Mov10l1 | ID: uc007xey.1_intron_11_0_chr15_88836198_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(1) OVARY |
(2) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
| GGTTGTCAGTGTACGTGTCACAGGGGTGTTGCTGATGTGAATGGAGACATCCTCCTGTTTGTATAGCTTAAGGATATGTAAGATACCAAACTTTGAAACAATATTTTCATATTGTATTGTCAAATACAGATGCGAGATTTTCCCAGGAAGGCCAGTCTACTTTGTAGAAGTTGAATCCCTCACTTATAATTTTTTTTTAGATTCATGAAGAAGATGTAACTCTTAAACTTAATCCAGGATTTGAACAAAT ......................................................................................................((((((((.(((((((....)))))))))).)))))......((..(((....((.(((((...))))).))...)))..)).................................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................AACTTTGAAACAATATTTTCATATTGTATTGTCAAATACAGATGCGAGATTT.............................................................................................................. | 52 | 1 | 31.00 | 31.00 | 31.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TTAAACTTAATCCAGGATTTGAACAAAT | 28 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................................................................................................................................ATGAAGAAGATGTAACTCTT.......................... | 20 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................GTAACTCTTAAACTTAATCCAGGATTTGAAC.... | 31 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TAAACTTAATCCAGGATTTGAACAAAT | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................TTGTATTGTCAAATACAGATGCGAGATTT.............................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGTCAAATACAGATGCGAGATTTTCCCA......................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................................................................................................CTTAAACTTAATCCAGGATTTGAACA... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAAGAAGATGTAACTCTTAAACTTcat................. | 28 | cat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................................................................................GAAGAAGATGTAACTCTTAAACTTAATCC............... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAAGAAGATGTAACTCTTAAACTTAATC................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................TGTCAAATACAGATGCGAGATTTTCCC.......................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TAAACTTAATCCAGGATTTGAACAAA. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAAGAAGATGTAACTCTTAAACTTAATCC............... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................ATCCAGGATTTGAACAA.. | 17 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| GGTTGTCAGTGTACGTGTCACAGGGGTGTTGCTGATGTGAATGGAGACATCCTCCTGTTTGTATAGCTTAAGGATATGTAAGATACCAAACTTTGAAACAATATTTTCATATTGTATTGTCAAATACAGATGCGAGATTTTCCCAGGAAGGCCAGTCTACTTTGTAGAAGTTGAATCCCTCACTTATAATTTTTTTTTAGATTCATGAAGAAGATGTAACTCTTAAACTTAATCCAGGATTTGAACAAAT ......................................................................................................((((((((.(((((((....)))))))))).)))))......((..(((....((.(((((...))))).))...)))..)).................................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................TGTTGCTGATGTGAacca.................................................................................................................................................................................................................. | 18 | acca | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |