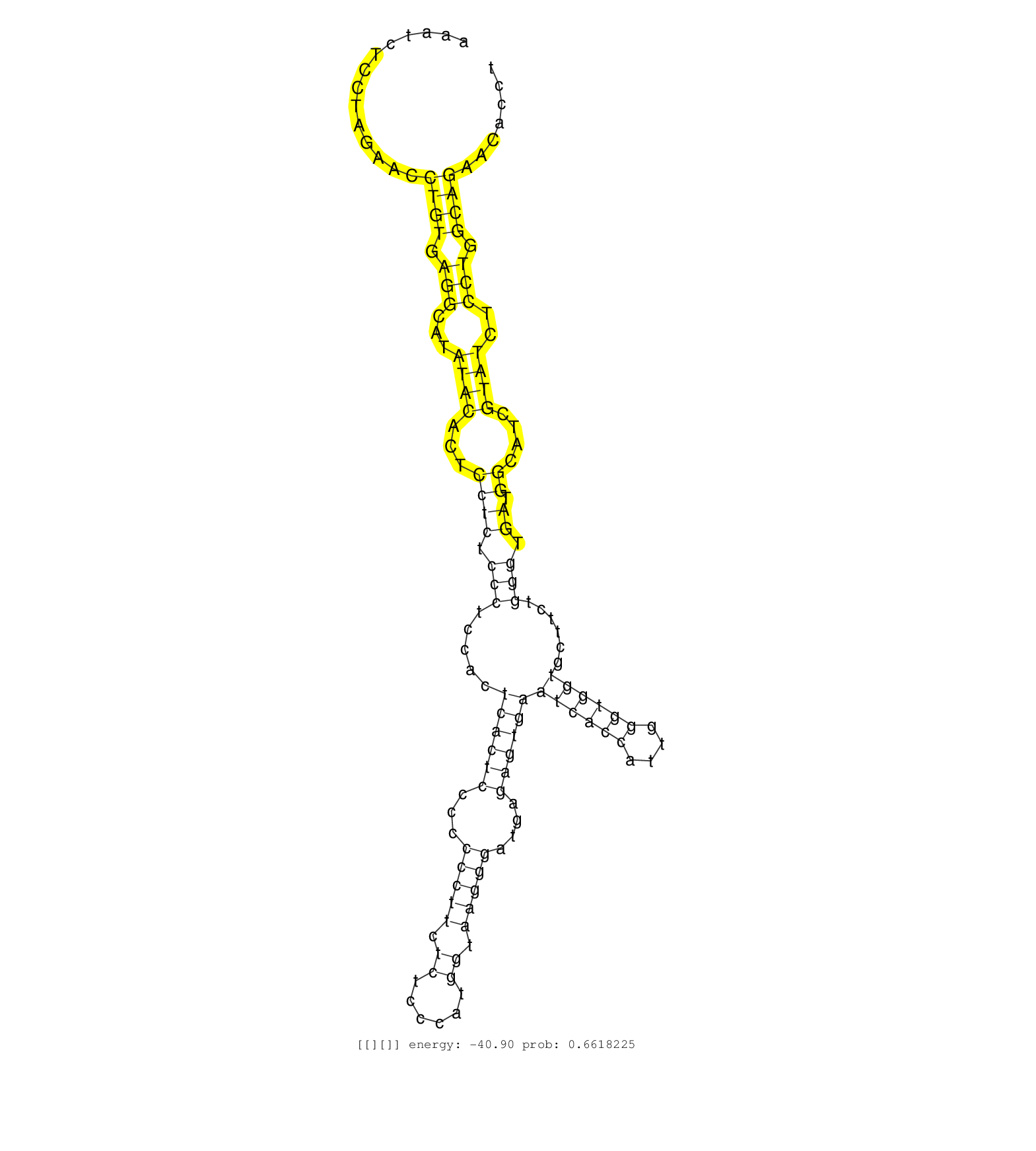

| Gene: Ribc2 | ID: uc007xcy.1_intron_2_0_chr15_84965990_f.3p | SPECIES: mm9 |
 |
 |
|
(6) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| AATATCATCGATCCATAGAGATTGCTTTAGTACCCCTTGCTCACAAAGATGTAGGAGCCACACTTAAATCTCCTAGAACCTGTGAGGCATATACACTCCTCTCCCTCCACTCACTCCCCCCCTTCTCTCCCATGGTAAGGGATGAGAGTGAATCACCATTGGGTGGTGCTTCTGGGTGATGGCATCGTATCTCCTGGCAGAACACCTCCACACTCAGACGAGACTCAAGTTTGATGAAACAGCCAGAGAG ...............................................................................((((.(((...((((...((((.(((.....((((((...(((((.((......)).)))))....))))))((((((....))))))......))).)).))....))))..))).)))).................................................. .................................................................66...........................................................................................................................................207......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................TCCTAGAACCTGTGAGGCATATACACTC........................................................................................................................................................ | 28 | 1 | 28.00 | 28.00 | 11.00 | 12.00 | 2.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................TCCTAGAACCTGTGAGGCATATACACTCC....................................................................................................................................................... | 29 | 1 | 8.00 | 8.00 | 3.00 | 2.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TGGCAGAACACCTCCACACTCAGACGAGAC.......................... | 30 | 1 | 5.00 | 5.00 | 2.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................TAGAACCTGTGAGGCATATACACTCCT...................................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | 1.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................TTAAATCTCCTAGAACCTGTGAGGCATA............................................................................................................................................................... | 28 | 1 | 4.00 | 4.00 | - | - | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................TCCTAGAACCTGTGAGGCATATACACT......................................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGGCATCGTATCTCCTGGCAGAACACCT........................................... | 28 | 1 | 3.00 | 3.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................GACGAGACTCAAGTTTGATGAAACAGCCAGA... | 31 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................................................................................GACGAGACTCAAGTTTGATGAAACAGCC...... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................TGGCAGAACACCTCCACACTCAGACGA............................. | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................CGAGACTCAAGTTTGATGAAACAGCCA..... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................TGGCAGAACACCTCCACACTCAGACGAGACT......................... | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TGGCAGAACACCTCCACACTCAGACGt............................. | 27 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................CAGACGAGACTCAAGTTTGATGAAACAGC....... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TAGAGATTGCTTTAGTACCCCTTGCTC................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGAACACCTCCACACTCAGACGAGACTC........................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGATGGCATCGTATCTCCTGGCAGAAC............................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................AAATCTCCTAGAACCTGTGAGGCATATA............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................CCTAGAACCTGTGAGGCATATACACTCC....................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................CGAGACTCAAGTTTGATGAAACAGCCAG.... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TTAAATCTCCTAGAACCTGTGAGGCAT................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CTCAGACGAGACTCAAGTT................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................................................................TTCTGGGTGATGGCATCGTATCTCCT....................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................CCTAGAACCTGTGAGGCATATACACTCCT...................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TCTCCTAGAACCTGTGAGGCATATACACTC........................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TAGAACCTGTGAGGCATATACACTCC....................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TGGCAGAACACCTCCACACTCAGACGAGA........................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................................................................................................................ACGAGACTCAAGTTTGATGAAACAGCCAGA... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................................................................................CGAGACTCAAGTTTGATGAAACAGCC...... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| AATATCATCGATCCATAGAGATTGCTTTAGTACCCCTTGCTCACAAAGATGTAGGAGCCACACTTAAATCTCCTAGAACCTGTGAGGCATATACACTCCTCTCCCTCCACTCACTCCCCCCCTTCTCTCCCATGGTAAGGGATGAGAGTGAATCACCATTGGGTGGTGCTTCTGGGTGATGGCATCGTATCTCCTGGCAGAACACCTCCACACTCAGACGAGACTCAAGTTTGATGAAACAGCCAGAGAG ...............................................................................((((.(((...((((...((((.(((.....((((((...(((((.((......)).)))))....))))))((((((....))))))......))).)).))....))))..))).)))).................................................. .................................................................66...........................................................................................................................................207......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................AGAACACCTCCACACTca.................................... | 18 | ca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |