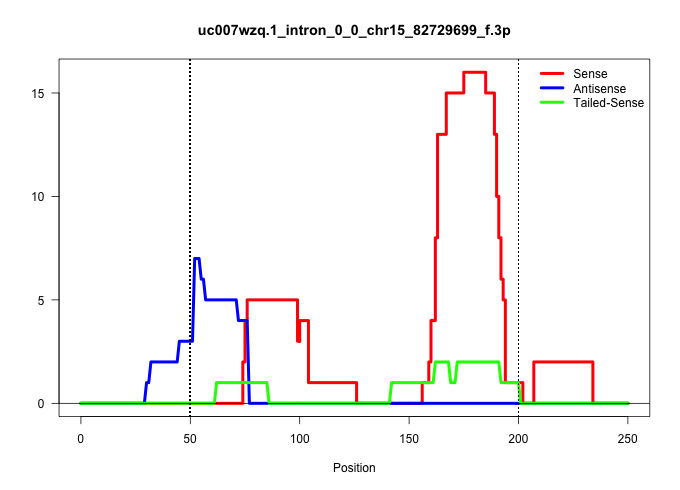
| Gene: AK080278 | ID: uc007wzq.1_intron_0_0_chr15_82729699_f.3p | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(14) TESTES |
| AAAGGGCTAAATTTCTTTTTCCTCAAGAAAAATCTCATGTACAACCACCGTCTCATAGATCAGTGGTTCTCAACCTTCCTAATGCTTTGACCCATTAATATAGTTCATATTGTGGTGACCCTAACCATAAACTATGTCAATGCTACTTCATAACTGTAATTTTGCTACTGTTATGAATAGTAATCTCTGTTTTCTGACAGCCTCTCCTGTGAAAGGGTCATTCACTACCAAAGCTGTCCCGACGCCTAGG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................GCTACTGTTATGAATAGTAATCTCTGTTTTC........................................................ | 31 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TGCTACTGTTATGAATAGTAATCTCTGTTT.......................................................... | 30 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TGCTACTGTTATGAATAGTAATCTCTGTT........................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................CTGTTATGAATAGTAATCTCTGTTTTC........................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGTGAAAGGGTCATTCACTACCAAAGC................ | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................................................................................................TTTGCTACTGTTATGAATAGTAATCTCTGT............................................................ | 30 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TTCCTAATGCTTTGACCCATTAATATAGT.................................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................CTTCCTAATGCTTTGACCCATTAAT....................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CTACTTCATAACTGTAATTTTGCTcaa................................................................................. | 27 | caa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TAGTTCATATTGTGGTGACCCTAACC............................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................GCTACTGTTATGAATAGTAATCTCTGT............................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................AATAGTAATCTCTGTTTTCTGACAGCC................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TCCTAATGCTTTGACCCATTAATATAGT.................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................GTGGTTCTCAACCTTCCTAAgggt.................................................................................................................................................................... | 24 | gggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................................TTTTGCTACTGTTATGAATAGTAATCTCTG............................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................GCTACTGTTATGAATAGTAATCTCTG............................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................ATGAATAGTAATCTCTGTTTTCTGACAGa................................................. | 29 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TGCTACTGTTATGAATAGTAATCTCTGTTa.......................................................... | 30 | a | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TAATTTTGCTACTGTTATGAATAGTAATC................................................................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................GCTACTGTTATGAATAGTAATCTCTGTTTT......................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| AAAGGGCTAAATTTCTTTTTCCTCAAGAAAAATCTCATGTACAACCACCGTCTCATAGATCAGTGGTTCTCAACCTTCCTAATGCTTTGACCCATTAATATAGTTCATATTGTGGTGACCCTAACCATAAACTATGTCAATGCTACTTCATAACTGTAATTTTGCTACTGTTATGAATAGTAATCTCTGTTTTCTGACAGCCTCTCCTGTGAAAGGGTCATTCACTACCAAAGCTGTCCCGACGCCTAGG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................TCATAGATCAGTGGTTCTCAACCTT............................................................................................................................................................................. | 25 | 1 | 4.00 | 4.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................GATCAGTGGTTCTCAACCTTCCaaag........................................................................................................................................................................... | 26 | aaag | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ...................................................TAGATCAGTGGTTCTCAACCTTCCggaa........................................................................................................................................................................... | 28 | ggaa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................AATCTCATGTACAACCACCGTCTCA................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................CACCGTCTCATAGATCAGTGGTTCTCA.................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................TCTCATGTACAACCACCGTCTCATA................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |