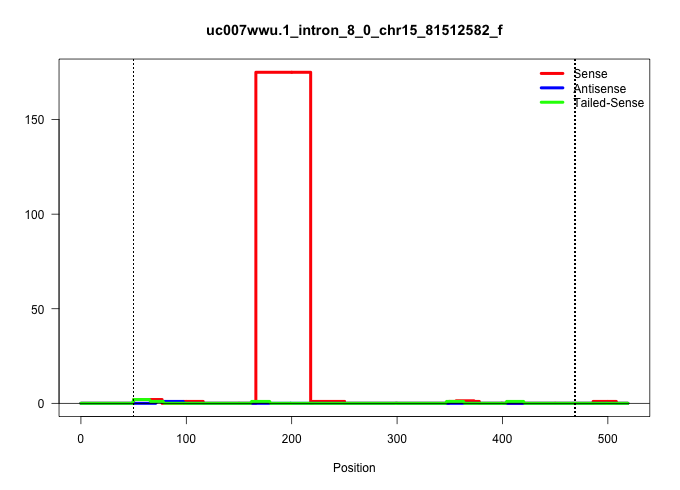
| Gene: L3mbtl2 | ID: uc007wwu.1_intron_8_0_chr15_81512582_f | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| CCACCCAGTGGGTTGGTCCCGGCGTGTTGGCCACGGCATCAAGATGTCAGGTCAGTAGAACCCCTGGCTGTTGCGACTCCAGGTTCAGCCTCCCCCTAAAGATCAGCAGGACCTTTGCTGTGCCCCACAGTAGTGCGATGAAGTGGCCTGTCATTGCTACCCTGCAAGTGACAATGTTGAGGATCAGAGAGGCCAGTGACTCAGGTCACTCAGCTCAGTATGAACAGCAAAACCAAGATGTAACACAGATGGTATTGCCACTGGCCCAGAACCAGAGAGCCTGGATCCCCCATGCTGCATAGTCCGCACTACTGCTCCAGGGTCCTCTGTTTTCCCATTTAGGGAAGCAGCTGGGGCACAACTGAGAACAAAGGTAGCCTGCCTGGTCACACACTTCTGGGAGAAGCATGGGATAACCAGGGGCTGCCTTCACCTCCCTTTTCCTCCATCTTCTCCGTCATCTCCCTAGACAGACGATGTGACATGTCTCATCACCCCACCTTCCGGAAAATCTACTGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................AGTGACAATGTTGAGGATCAGAGAGGCCAGTGACTCAGGTCACTCAGCTCAG............................................................................................................................................................................................................................................................................................................. | 52 | 1 | 175.00 | 175.00 | 175.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTCAGTAGAACCCCTGGCTGTTGCGAC.......................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................................................................................................................................................TGAACAGCAAAACCAAGATGTAACACAGAT............................................................................................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................GCTGGGGCACAACTGAGAACAAAGGTAGC............................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCTCATCACCCCACCTTCCGGA........... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................................................................CAGCTGGGGCACAACTc........................................................................................................................................................... | 17 | c | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TGCAAGTGACAATGcaag................................................................................................................................................................................................................................................................................................................................................... | 18 | caag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................AGCATGGGATAACatca.................................................................................................. | 17 | atca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTCAGTAGAACCCCTGGCTGTTGCGgctt........................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 29 | gctt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................AAAGATCAGCAGGACCTTT................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTCAGTAGAACCgagg..................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 16 | gagg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................................................................................CACAACTGAGAACAAAG.................................................................................................................................................. | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| CCACCCAGTGGGTTGGTCCCGGCGTGTTGGCCACGGCATCAAGATGTCAGGTCAGTAGAACCCCTGGCTGTTGCGACTCCAGGTTCAGCCTCCCCCTAAAGATCAGCAGGACCTTTGCTGTGCCCCACAGTAGTGCGATGAAGTGGCCTGTCATTGCTACCCTGCAAGTGACAATGTTGAGGATCAGAGAGGCCAGTGACTCAGGTCACTCAGCTCAGTATGAACAGCAAAACCAAGATGTAACACAGATGGTATTGCCACTGGCCCAGAACCAGAGAGCCTGGATCCCCCATGCTGCATAGTCCGCACTACTGCTCCAGGGTCCTCTGTTTTCCCATTTAGGGAAGCAGCTGGGGCACAACTGAGAACAAAGGTAGCCTGCCTGGTCACACACTTCTGGGAGAAGCATGGGATAACCAGGGGCTGCCTTCACCTCCCTTTTCCTCCATCTTCTCCGTCATCTCCCTAGACAGACGATGTGACATGTCTCATCACCCCACCTTCCGGAAAATCTACTGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................GCGACTCCAGGTTCAGCCTCCCCCTA..................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |