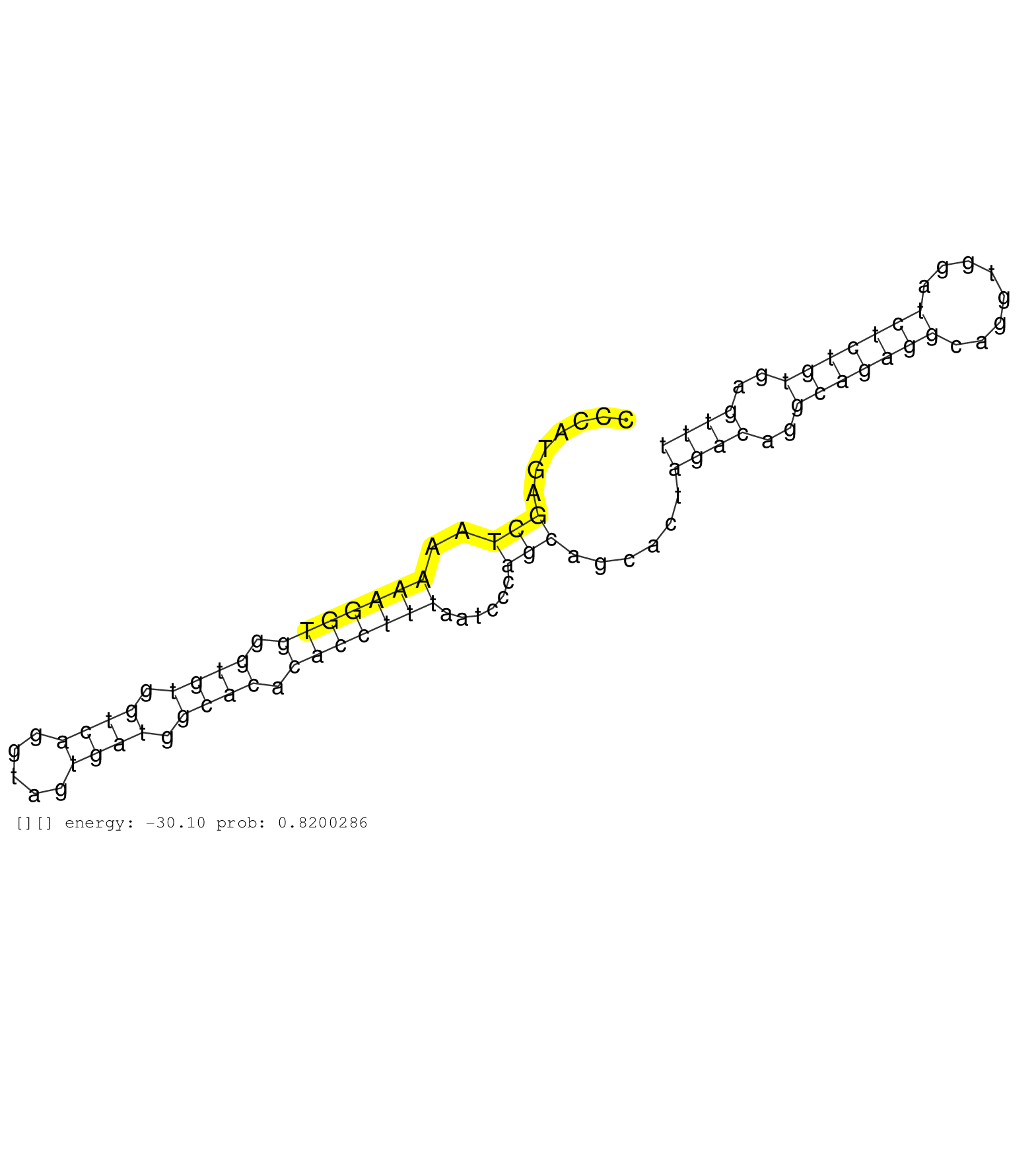

| Gene: St13 | ID: uc007wwl.1_intron_4_0_chr15_81205337_r.5p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(3) PIWI.ip |
(15) TESTES |
| ATAAACCCTGATTCCGCTCAGCCATACAAATGGCGAGGGAAAGCACACAGGTAAATGAGCAAATAAATACTGCTTAGAGAGCTGAGTTTGACCGGTAGAACCCATGAGCTAAAAAGGTGGGTGTGGTCAGGTAGTGATGGCACACACCTTTAATCCCAGCAGCACTAGACAGGCAGAGGCAGGTGGATCTCTGTGAGTTTGAGACCAGCATGGAATCTAGAGAGCTCAGGACAACCAGGGCTACAGATAT ...........................................................................................................(((..(((((((.((((.((((.....)))).)))).)))))))......)))......((((..(((((((........)))))))..)))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................GAGCAAATAAATACTGCTTAGAGAGCTGAGTTTGACCGGTAGAACCCATGAG.............................................................................................................................................. | 52 | 1 | 33.00 | 33.00 | 33.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................CCATACAAATGGCGAGGGAAAGCACA........................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................GAGGGAAAGCACACtata...................................................................................................................................................................................................... | 18 | tata | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................CTAGAGAGCTCAGGACAACCAG............ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................TACAAATGGCGAGGGAAAGCACACAG........................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................ATACAAATGGCGAGGGAAAGCACACAGactc.................................................................................................................................................................................................... | 31 | actc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................................................GCTCAGGACAACCAGtccc........ | 19 | tccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ATAAACCCTGATTCCGCTCAGCCATACAAATGGCGAGGGAAAGCACACAGGTAAATGAGCAAATAAATACTGCTTAGAGAGCTGAGTTTGACCGGTAGAACCCATGAGCTAAAAAGGTGGGTGTGGTCAGGTAGTGATGGCACACACCTTTAATCCCAGCAGCACTAGACAGGCAGAGGCAGGTGGATCTCTGTGAGTTTGAGACCAGCATGGAATCTAGAGAGCTCAGGACAACCAGGGCTACAGATAT ...........................................................................................................(((..(((((((.((((.((((.....)))).)))).)))))))......)))......((((..(((((((........)))))))..)))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................GTGATGGCACACACCTTTAATCCCAatt............................................................................................ | 28 | att | 2.00 | 0.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................GTGATGGCACACACCTTTAATCCCAaaaa............................................................................................ | 29 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................CTCAGGACAACCAGGGCTACAaca..... | 24 | aca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................................GTGATGGCACACACCTTTAATCCCAcgta............................................................................................ | 29 | cgta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................AGTGATGGCACACACCTTTAATCCCAgtaa............................................................................................ | 30 | gtaa | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................GATGGCACACACCTTTAATCCCAgata............................................................................................ | 27 | gata | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................GTGATGGCACACACCTTTAATCCCAatat............................................................................................ | 29 | atat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................ATGGCACACACCTTTAATCCCAaca............................................................................................ | 25 | aca | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TGATGGCACACACCTTTAATCCCAaatc............................................................................................ | 28 | aatc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................GTGATGGCACACACCTTTAATCCCAgca............................................................................................ | 28 | gca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................TAGTGATGGCACACACCTTTAATCCCAtcct............................................................................................ | 31 | tcct | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................TAGTGATGGCACACACCTTTAATCCCAa............................................................................................ | 28 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................GATGGCACACACCTTTAATCCCAgggt............................................................................................ | 27 | gggt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................GTGATGGCACACACCTTTAATCCCAaga............................................................................................ | 28 | aga | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TGATGGCACACACCTTTAATCCCAaat............................................................................................ | 27 | aat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................GATGGCACACACCTTTAATCCCAggt............................................................................................ | 26 | ggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................................TGATGGCACACACCTTTAATCCCAGCagaa.......................................................................................... | 30 | agaa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |