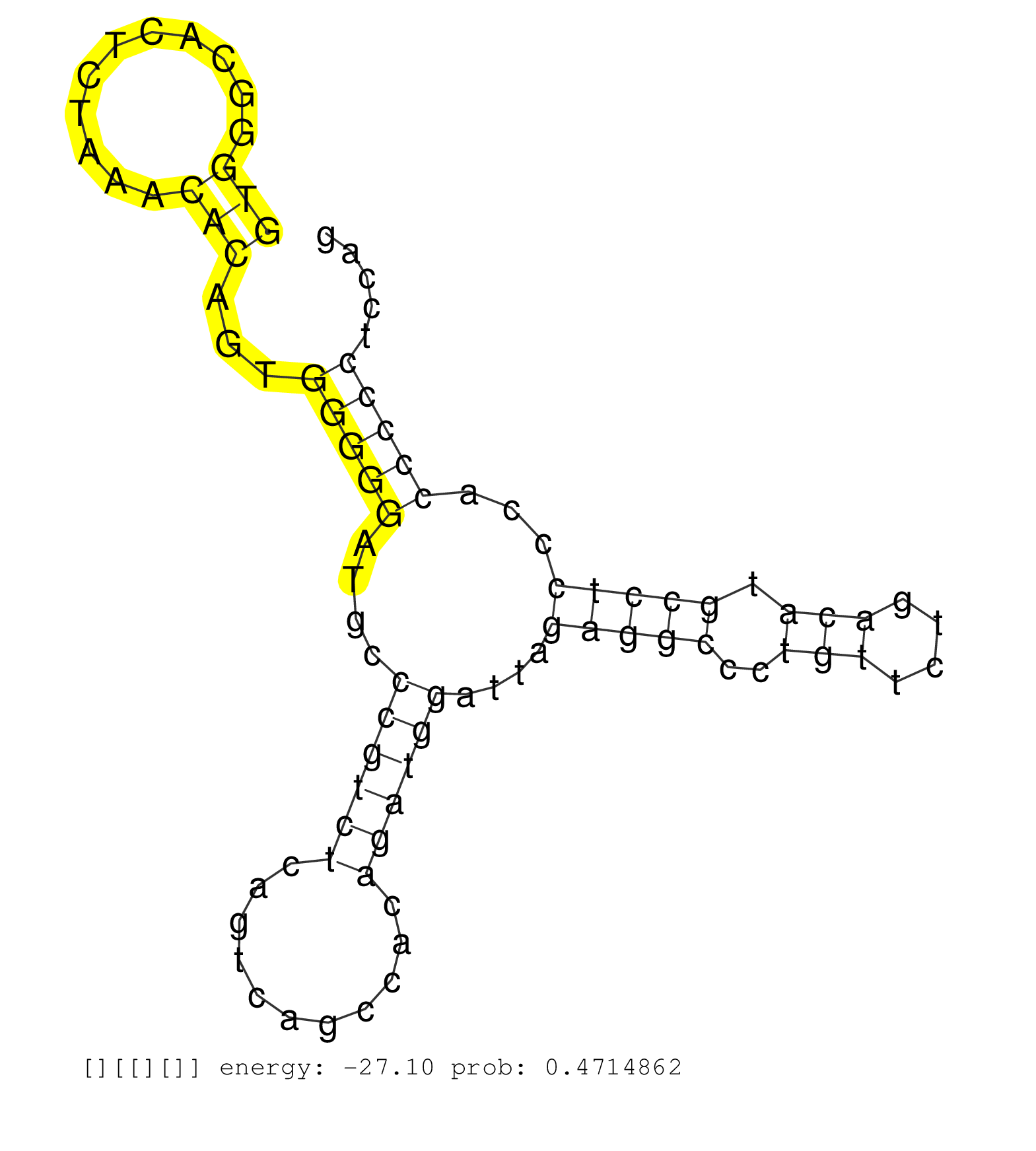
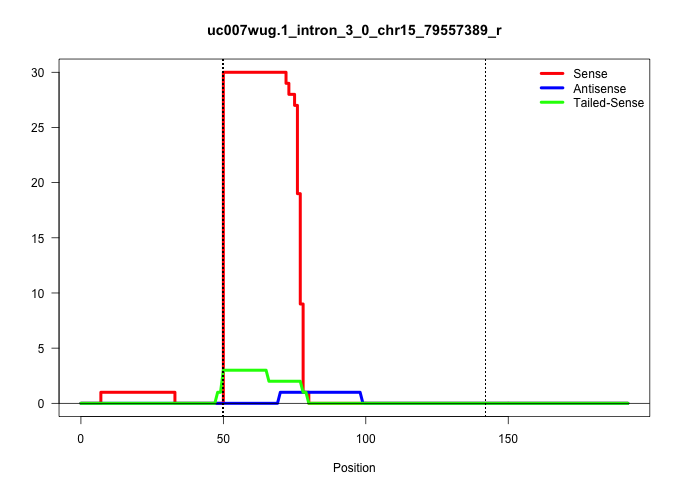
| Gene: Unc84b | ID: uc007wug.1_intron_3_0_chr15_79557389_r | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(3) PIWI.mut |
(18) TESTES |
| TTGGCATCCCCCTGTGGTACCACTCCCAGTCACCTCGGGTCATTCTGCAGGTGGGCACTCTAAACACAGTGGGGGATGCCCGTCTCAGTCAGCCACAGATGGATTAGAGGCCCTGTTCTGACATGCCTCCCACCCCCTCCAGCCAGATGTGCACCCAGGCAACTGCTGGGCCTTCCAGGGGCCCCAGGGCTT ..................................................(((...........)))...(((((....((((((...........))))))....(((((..(((....))).)))))...)))))....................................................... ..................................................51.........................................................................................142................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGGGCACTCTAAACACAGTGGGGGA.................................................................................................................... | 26 | 1 | 11.00 | 11.00 | - | 3.00 | 2.00 | - | - | 3.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTGGGCACTCTAAACACAGTGGGGGAT................................................................................................................... | 27 | 1 | 10.00 | 10.00 | - | 3.00 | 1.00 | - | 3.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTGGGCACTCTAAACACAGTGGGGGATG.................................................................................................................. | 28 | 1 | 8.00 | 8.00 | 5.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTGGGCACTCTAAACACAGTGGGGG..................................................................................................................... | 25 | 1 | 6.00 | 6.00 | - | - | - | 4.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ..................................................GTGGGCACTCTAAACACAGTGGG....................................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................AGGTGGGCACTCTAAgga.............................................................................................................................. | 18 | gga | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......CCCCCTGTGGTACCACTCCCAGTCAC............................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCACTCTAAACACAGTGGGGGATGCC................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCACTCTAAACACAGTGGGGGATGC................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTGGGCACTCTAAACACAGTGGGGGATa.................................................................................................................. | 28 | a | 1.00 | 10.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGCACTCTAAACACAGTGG........................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTGGGCACTCTAAACACAGTGGGGGATttt................................................................................................................ | 30 | ttt | 1.00 | 10.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................CAGGTGGGCACTCTAAACACAGTGGGGGAT................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| TTGGCATCCCCCTGTGGTACCACTCCCAGTCACCTCGGGTCATTCTGCAGGTGGGCACTCTAAACACAGTGGGGGATGCCCGTCTCAGTCAGCCACAGATGGATTAGAGGCCCTGTTCTGACATGCCTCCCACCCCCTCCAGCCAGATGTGCACCCAGGCAACTGCTGGGCCTTCCAGGGGCCCCAGGGCTT ..................................................(((...........)))...(((((....((((((...........))))))....(((((..(((....))).)))))...)))))....................................................... ..................................................51.........................................................................................142................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................GGGGGATGCCCGTCTCAGTCAGCCACAGA............................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TGGGGGATGCCCGTCTCAGTCAGCCAa................................................................................................. | 27 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |