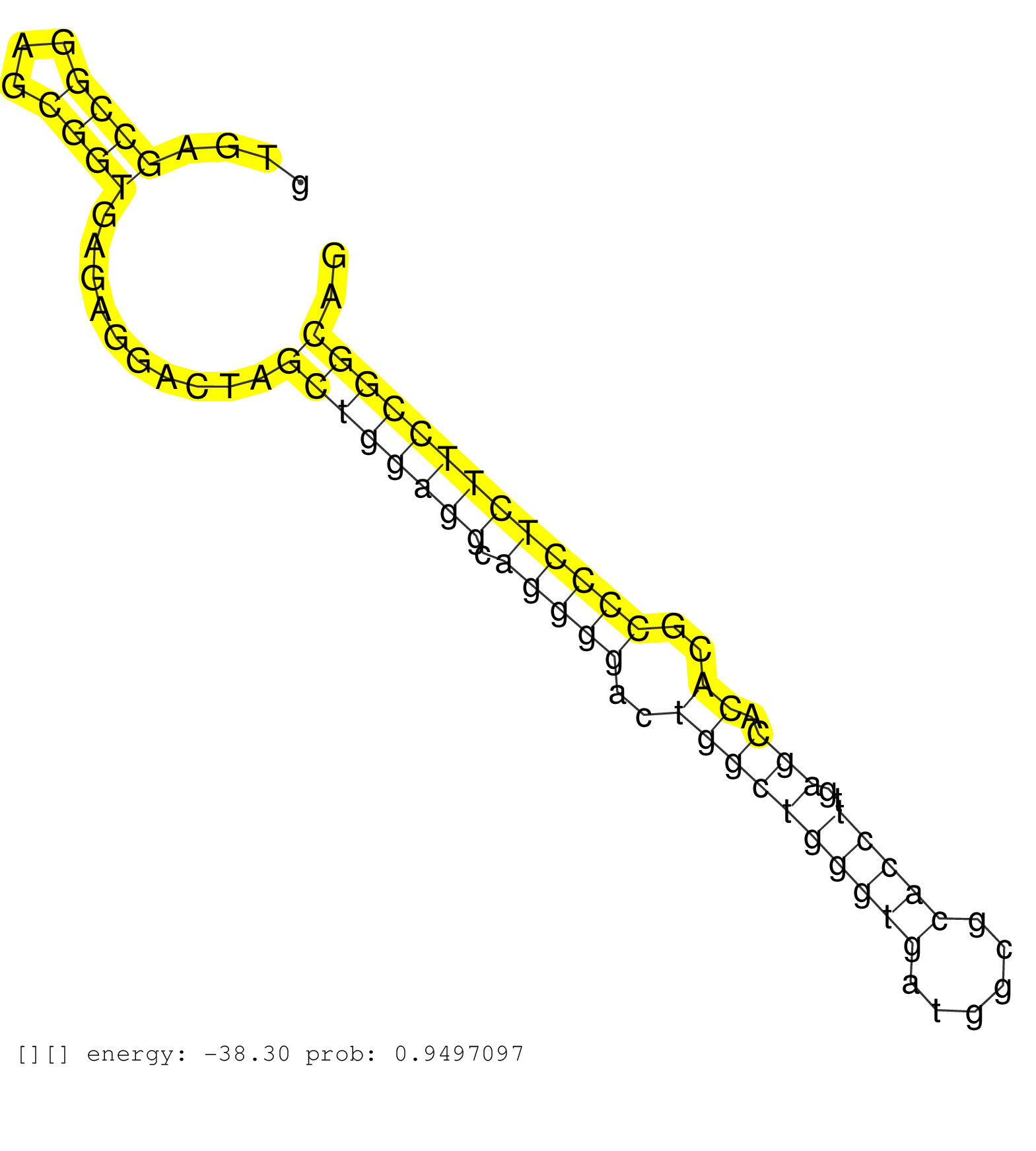
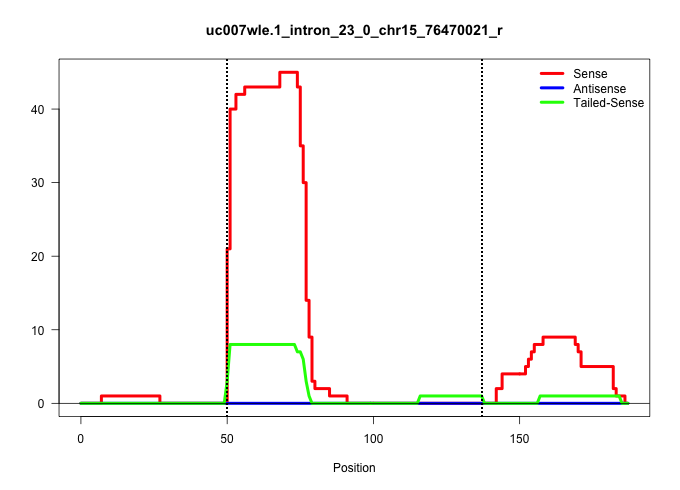
| Gene: Nfkbil2 | ID: uc007wle.1_intron_23_0_chr15_76470021_r | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(12) TESTES |
| CGAGGAGGCGGCCTACTGCCACCAGCTGGGGGAGCTGCTGGCTAGCCACGGTGAGCCGGAGCGGTGAGAGGACTAGCTGGAGGCAGGGGACTGGCTGGGTGATGGCGCACCTTGAGCACACGCCCCTCTTCCGGCAGGCCGCTTCAAGGACGCTTTGGAGGAGCACCAGCAGGAACTACATCTGCTA ......................................................((((...))))..........((((((((.(((((..((((((((((......)))))..))).))..))))))))))))).................................................... ..................................................51....................................................................................137................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................TGAGCCGGAGCGGTGAGAGGACTAGC.............................................................................................................. | 26 | 1 | 9.00 | 9.00 | - | 5.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTGAGCCGGAGCGGTGAGAGGACTA................................................................................................................ | 25 | 1 | 8.00 | 8.00 | 3.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 |
| ..................................................GTGAGCCGGAGCGGTGAGAGGACTAGC.............................................................................................................. | 27 | 1 | 7.00 | 7.00 | 5.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................TGAGCCGGAGCGGTGAGAGGACTAGCTG............................................................................................................ | 28 | 1 | 4.00 | 4.00 | 1.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - |
| ...................................................TGAGCCGGAGCGGTGAGAGGACTAGCT............................................................................................................. | 27 | 1 | 4.00 | 4.00 | 1.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCCGGAGCGGTGAGAGGACTAG............................................................................................................... | 26 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGAGCCGGAGCGGTGAGAGGACT................................................................................................................. | 24 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TTTGGAGGAGCACCAGCAGGAACTACA....... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - |
| ................................................................................................................................................CAAGGACGCTTTGGAGGAGCACCAGCA................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - |
| .....................................................AGCCGGAGCGGTGAGAGGACTAGCTG............................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................TGAGCCGGAGCGGTGAGAGGACTAG............................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TTCAAGGACGCTTTGGAGGAGCACCAG.................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTGAGCCGGAGCGGTGAGAGGACTAGCT............................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................TTCAAGGACGCTTTGGAGGAGCACCAGC................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................TGAGCCGGAGCGGTGAGAGGACTAGCTt............................................................................................................ | 28 | t | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................AGGACTAGCTGGAGGCA...................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCCGGAGCGGTGAGAGGgct................................................................................................................. | 24 | gct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................CACACGCCCCTCTTCCGGCAGt................................................. | 22 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................TGAGCCGGAGCGGTGAGAGGACTggct............................................................................................................. | 27 | ggct | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................AGGACTAGCTGGAGGCAGGGGAC................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................CGGAGCGGTGAGAGGACTAGCTGG........................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......GCGGCCTACTGCCACCAGCT................................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................................TTTGGAGGAGCACCAGCAGGAACTACATC..... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................AGGAGCACCAGCAGGAACTACATCTGCT. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................TGAGCCGGAGCGGTGAGAGGACTAGCc............................................................................................................. | 27 | c | 1.00 | 9.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................CTTTGGAGGAGCACCAGCAGGAACTACATC..... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TTGGAGGAGCACCAGCAGGAACTACATC..... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGCCGGAGCGGTGAGAGGACTAGa.............................................................................................................. | 26 | a | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGGAGGAGCACCAGCAGGAACTACATCT.... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCCGGAGCGGTGAGAGGACTAaa.............................................................................................................. | 27 | aa | 1.00 | 8.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................GAGGAGCACCAGCAGGAACTACATCTGt.. | 28 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGCCGGAGCGGTGAGAGGACTAt............................................................................................................... | 25 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCCGGAGCGGTGAGAGGACTAGt.............................................................................................................. | 27 | t | 1.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| CGAGGAGGCGGCCTACTGCCACCAGCTGGGGGAGCTGCTGGCTAGCCACGGTGAGCCGGAGCGGTGAGAGGACTAGCTGGAGGCAGGGGACTGGCTGGGTGATGGCGCACCTTGAGCACACGCCCCTCTTCCGGCAGGCCGCTTCAAGGACGCTTTGGAGGAGCACCAGCAGGAACTACATCTGCTA ......................................................((((...))))..........((((((((.(((((..((((((((((......)))))..))).))..))))))))))))).................................................... ..................................................51....................................................................................137................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|