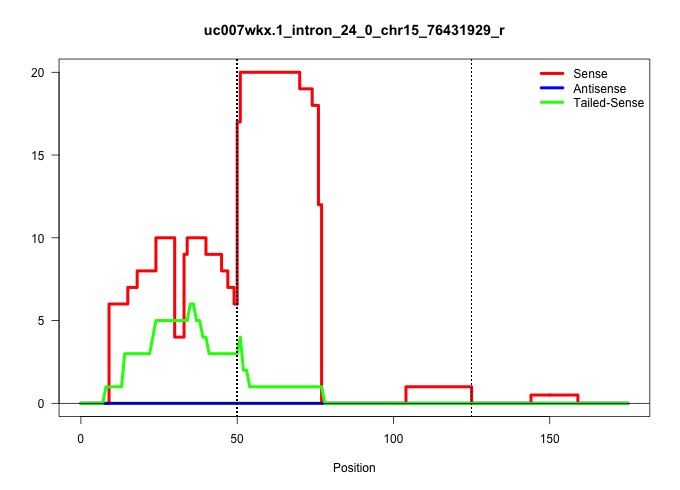
| Gene: Cpsf1 | ID: uc007wkx.1_intron_24_0_chr15_76431929_r | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(15) TESTES |
| AGAGCCACCCTCAAAGAAGAAGCGAGTGGAGCCTGCAGTGGGCTGGACAGGTGAGAGGCTGGACTAGAACGAGCAGGGGAGGCTAGCGTCCATAGTTGGACTCAGTTGGACTCTGTCTGCTACAGGAGGCAAGACGGTGCCACAGGACGAGGTGGATGAAATTGAAGTTTATGGC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGAGGCTGGACTAGAACGAGCAGG.................................................................................................. | 27 | 1 | 9.00 | 9.00 | 2.00 | - | - | 4.00 | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - |
| ..................................................GTGAGAGGCTGGACTAGAACGAGCAG................................................................................................... | 26 | 1 | 6.00 | 6.00 | 3.00 | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| .........CTCAAAGAAGAAGCGAGTGGA................................................................................................................................................. | 21 | 1 | 6.00 | 6.00 | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TGCAGTGGGCTGGACAG............................................................................................................................. | 17 | 1 | 5.00 | 5.00 | 1.00 | - | 1.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................TGAGAGGCTGGACTAGAACGAGCAGG.................................................................................................. | 26 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................GAGTGGAGCCTGCAGTGGGCTGGACAGGagg......................................................................................................................... | 31 | agg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAGGCTGGACTAGAACGAGC..................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............AGAAGAAGCGAGTGGAGCCTGCAGTGt...................................................................................................................................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................CAGTGGGCTGGACAGGa........................................................................................................................... | 17 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................GAAGCGAGTGGAGCCTGCAGTGGGCTG.................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................TGAGAGGCTGGACTAGAACGAGCAGGa................................................................................................. | 27 | a | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............GAAGAAGCGAGTGGAGCCTGCAGTG....................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................AGTGGAGCCTGCAGTGGGCTGGA................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........CCTCAAAGAAGAAGCGAGTGGAGCCTGCt.......................................................................................................................................... | 29 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................AGTGGAGCCTGCAGTGGGCTGGACA.............................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................GCAGTGGGCTGGACAG............................................................................................................................. | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................AGTGGAGCCTGCAGTGGGCTGGACAGGa........................................................................................................................... | 28 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTGAGAGGCTGGACTAGAAC......................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................GTTGGACTCTGTCTGCTACAG.................................................. | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............AGAAGAAGCGAGTGGAGCCTGtatt........................................................................................................................................ | 25 | tatt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................GGACGAGGTGGATGA................ | 15 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGAGCCACCCTCAAAGAAGAAGCGAGTGGAGCCTGCAGTGGGCTGGACAGGTGAGAGGCTGGACTAGAACGAGCAGGGGAGGCTAGCGTCCATAGTTGGACTCAGTTGGACTCTGTCTGCTACAGGAGGCAAGACGGTGCCACAGGACGAGGTGGATGAAATTGAAGTTTATGGC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|