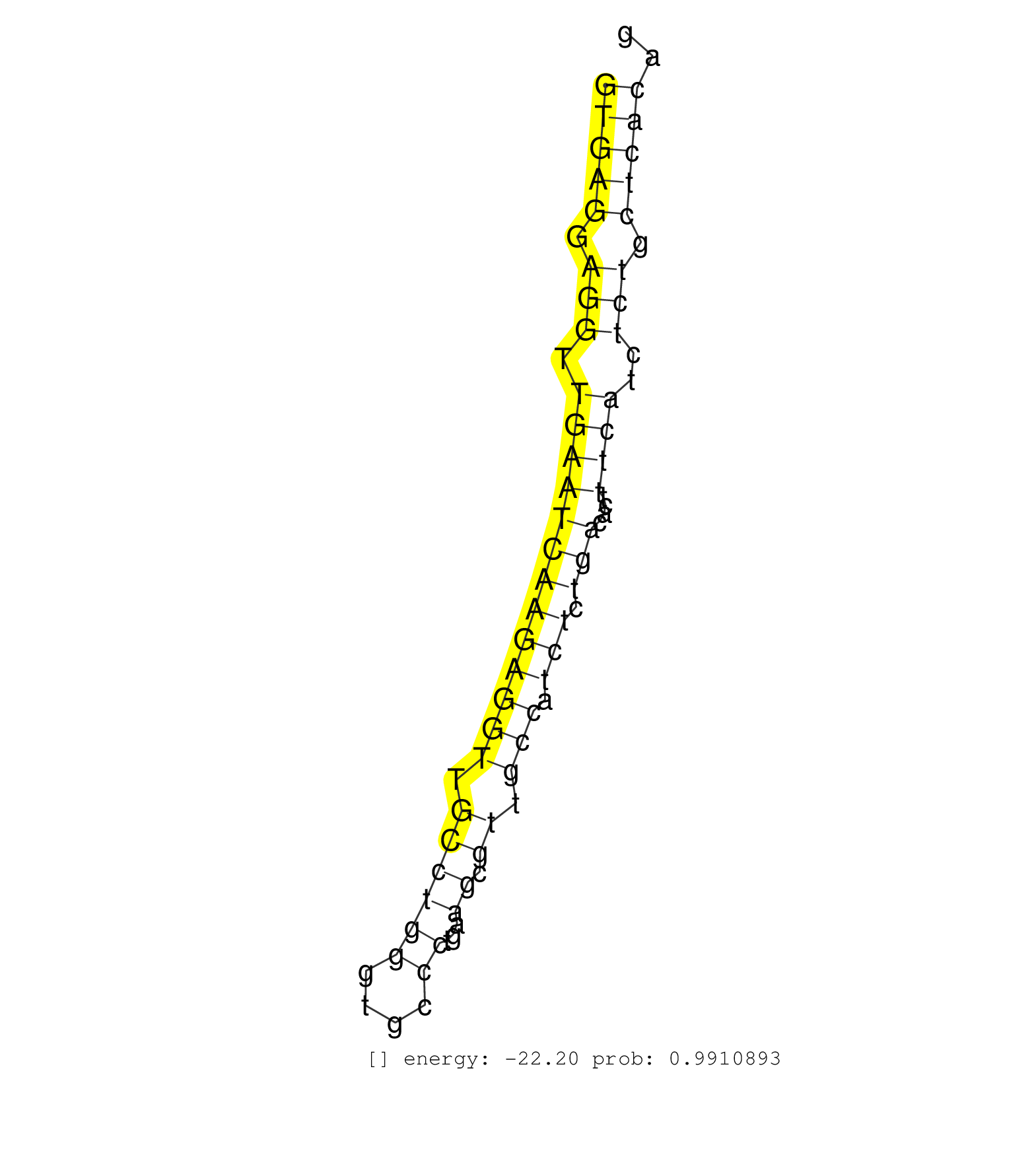
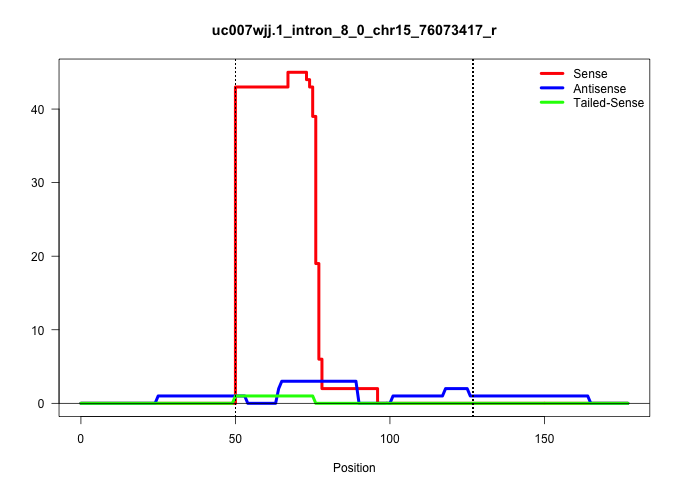
| Gene: Plec1 | ID: uc007wjj.1_intron_8_0_chr15_76073417_r | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(20) TESTES |
| CTGGCAGAGACTTGGCTGTGGGGGTGTCCTCATCTTTCAAGATCCTGCAGGTGAGGAGGTTGAATCAAGAGGTTGCCTGGGTGCCCTGAAGCGTTGCCATCTCTGACACTTTCATCTCTGCTCACAGATGCAAAGAGGGTCTTAGCCCAGGCAGAGCACCGACTACATGGTGTCCGA ..................................................(((((.(((.(((((((((((((.((((((....))...)).)).))).))).)))....))))..))).))))).................................................... ..................................................51..........................................................................127................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGGAGGTTGAATCAAGAGGTTGC..................................................................................................... | 26 | 1 | 20.00 | 20.00 | 4.00 | 3.00 | 4.00 | 1.00 | 2.00 | - | 1.00 | - | - | 2.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - |
| ..................................................GTGAGGAGGTTGAATCAAGAGGTTGCC.................................................................................................... | 27 | 1 | 13.00 | 13.00 | 4.00 | 2.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTGAGGAGGTTGAATCAAGAGGTTG...................................................................................................... | 25 | 1 | 4.00 | 4.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTGAGGAGGTTGAATCAAGAGGTTGCCT................................................................................................... | 28 | 1 | 4.00 | 4.00 | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................AGAGGTTGCCTGGGTGCCCTGAAGCGTTG................................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGAGGTTGAATCAAGAGGT........................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGAGGTTGAATCAAGAGGgtgc..................................................................................................... | 26 | gtgc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGAGGTTGAATCAAGAGGTT....................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTGGCAGAGACTTGGCTGTGGGGGTGTCCTCATCTTTCAAGATCCTGCAGGTGAGGAGGTTGAATCAAGAGGTTGCCTGGGTGCCCTGAAGCGTTGCCATCTCTGACACTTTCATCTCTGCTCACAGATGCAAAGAGGGTCTTAGCCCAGGCAGAGCACCGACTACATGGTGTCCGA ..................................................(((((.(((.(((((((((((((.((((((....))...)).)).))).))).)))....))))..))).))))).................................................... ..................................................51..........................................................................127................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................TCAAGAGGTTGCCTGGGTGCCCTGAA....................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................GCCCAGGCAGAGCACCGACTACA.......... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................GTCCTCATCTTTCAAGATCCTGCAGGTGA........................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................GTGTCCTCATCTTTCAAGATCCTGCA................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................TGCTCACAGATGCAAAGAGGGTCTTA................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................CAAGAGGTTGCCTGGGTGCCCTGAA....................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................GCCCAGGCAGAGCACCGACTA............ | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TCTGACACTTTCATCTCTGCTCACA................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGCCCAGGCAGAGCACCGACTACAcaga.......... | 28 | caga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |