
| Gene: Plec1 | ID: uc007wir.1_intron_9_0_chr15_76016468_r | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(6) PIWI.ip |
(2) PIWI.mut |
(27) TESTES |
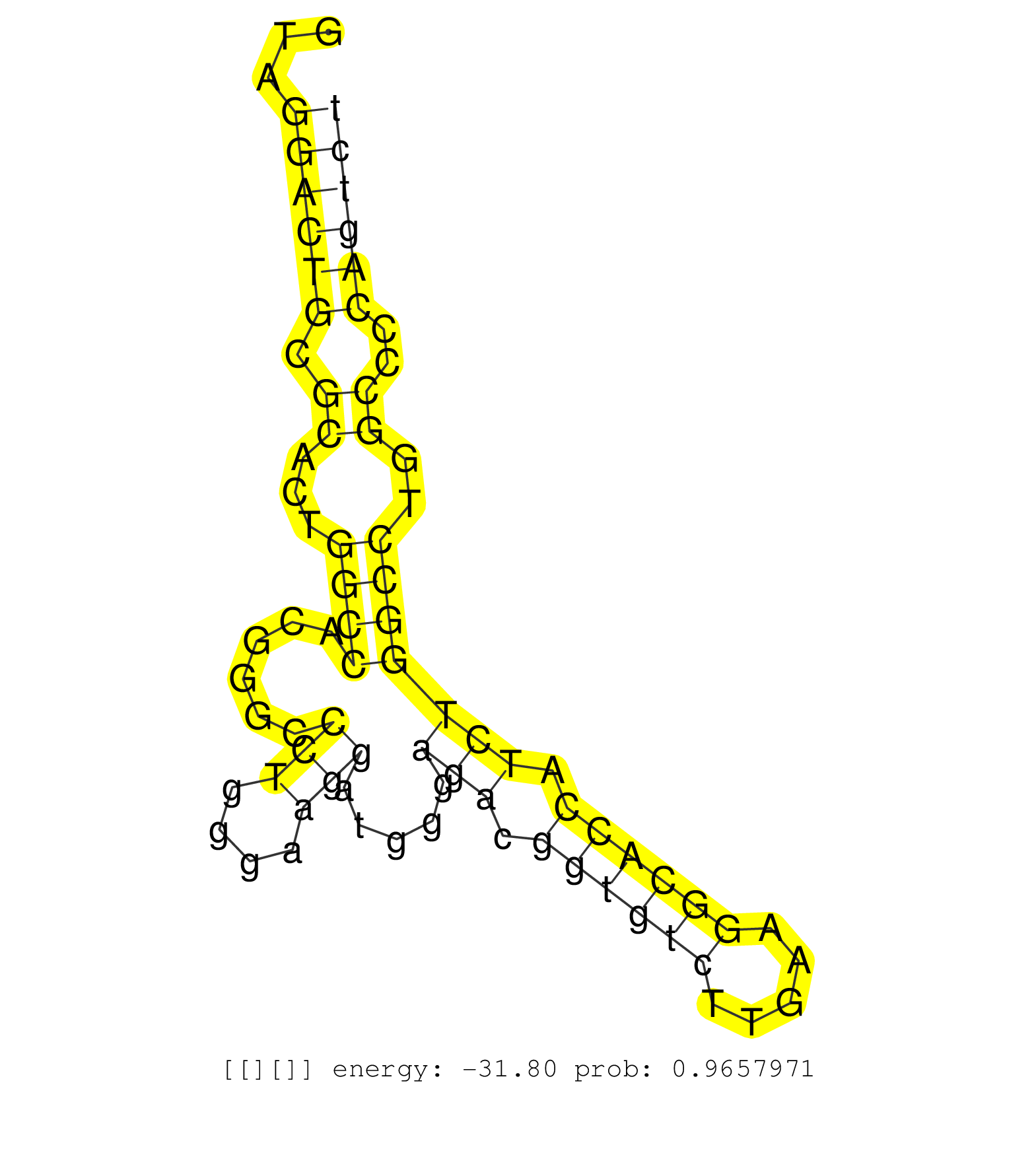
| AGAGCCTCAGTCGGGACATACAGCTCATCCGGTCCTGGTCCCTAGTCACGGTAGGACTGCGCACTGGCCACGGGCCCTGGGAAGGATGGGAGACGGTGTCTTGAAGGCACCATCTGGCCTGGCCCCAGTCTCAGTGACATGTGCCACCTCCAGTTCCGCACACTGAAGCCCGAGGAGCAGCGACAAGCTCTGCGCAACCTGGA .....................................................((((((.((...((((......(((....))).....(((.((((((.....)))))).)))))))..))..))))))........................................................................ ..................................................51..............................................................................131...................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGACTGCGCACTGGCCACGGGCCCT............................................................................................................................. | 28 | 1 | 34.00 | 34.00 | 3.00 | 2.00 | 5.00 | - | 4.00 | - | 3.00 | - | 4.00 | 2.00 | - | 2.00 | - | 2.00 | - | - | - | 1.00 | 1.00 | 1.00 | 2.00 | - | 1.00 | - | - | 1.00 | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGGCCC.............................................................................................................................. | 27 | 1 | 18.00 | 18.00 | 4.00 | 1.00 | 1.00 | - | - | 1.00 | 2.00 | 5.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGGC................................................................................................................................ | 25 | 1 | 8.00 | 8.00 | - | 1.00 | - | - | 1.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGGCCCTGGG.......................................................................................................................... | 31 | 1 | 7.00 | 7.00 | - | - | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGG.................................................................................................................................. | 23 | 1 | 6.00 | 6.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGGCC............................................................................................................................... | 26 | 1 | 4.00 | 4.00 | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGACTGCGCACTGGCCACGGGCCCTGG........................................................................................................................... | 29 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGGCa............................................................................................................................... | 26 | a | 3.00 | 8.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGACTGCGCACTGGCCACGGGCCCT............................................................................................................................. | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGG................................................................................................................................. | 24 | 1 | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGGCCCc............................................................................................................................. | 28 | c | 2.00 | 18.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGccct.............................................................................................................................. | 27 | ccct | 2.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .......CAGTCGGGACATACAGC................................................................................................................................................................................... | 17 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGGCCCa............................................................................................................................. | 28 | a | 2.00 | 18.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGACTGCGCACTGGCCACGGGCCC.............................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGa................................................................................................................................. | 24 | a | 1.00 | 6.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACt................................................................................................................................... | 22 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGGaa............................................................................................................................... | 26 | aa | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................CCGCACACTGAAGCCCGAGGAGCAGCG..................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................AGCTCATCCGGTCCTGGTCCCTAGTC............................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................ATACAGCTCATCCGGTCCTGGTCCCT................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................TAGGACTGCGCACTGGCCACGGGCCCTG............................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........GTCGGGACATACAGCTCATCC............................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TTGAAGGCACCATCTGGCCTGGCCCCA............................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TGGCCACGGGCCCTGGGAAGGATGGta................................................................................................................ | 27 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACG................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGACTGCGCACTGGCCACGG.................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGGCaa.............................................................................................................................. | 27 | aa | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGGCCCTa............................................................................................................................ | 29 | a | 1.00 | 34.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGGCCCTGtttt........................................................................................................................ | 33 | tttt | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGGCact............................................................................................................................. | 28 | act | 1.00 | 8.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGG........................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGACTGCGCACTGGCCACGGGCCCTG............................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TCCGGTCCTGGTCCCTAGTCACGttc...................................................................................................................................................... | 26 | ttc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| AGAGCCTCAGTCGGGACATACAGCTCATCCGGTCCTGGTCCCTAGTCACGGTAGGACTGCGCACTGGCCACGGGCCCTGGGAAGGATGGGAGACGGTGTCTTGAAGGCACCATCTGGCCTGGCCCCAGTCTCAGTGACATGTGCCACCTCCAGTTCCGCACACTGAAGCCCGAGGAGCAGCGACAAGCTCTGCGCAACCTGGA .....................................................((((((.((...((((......(((....))).....(((.((((((.....)))))).)))))))..))..))))))........................................................................ ..................................................51..............................................................................131...................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............CATACAGCTCATCCGgca............................................................................................................................................................................ | 18 | gca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................CCGGTCCTGGTCCCT................................................................................................................................................................ | 15 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |