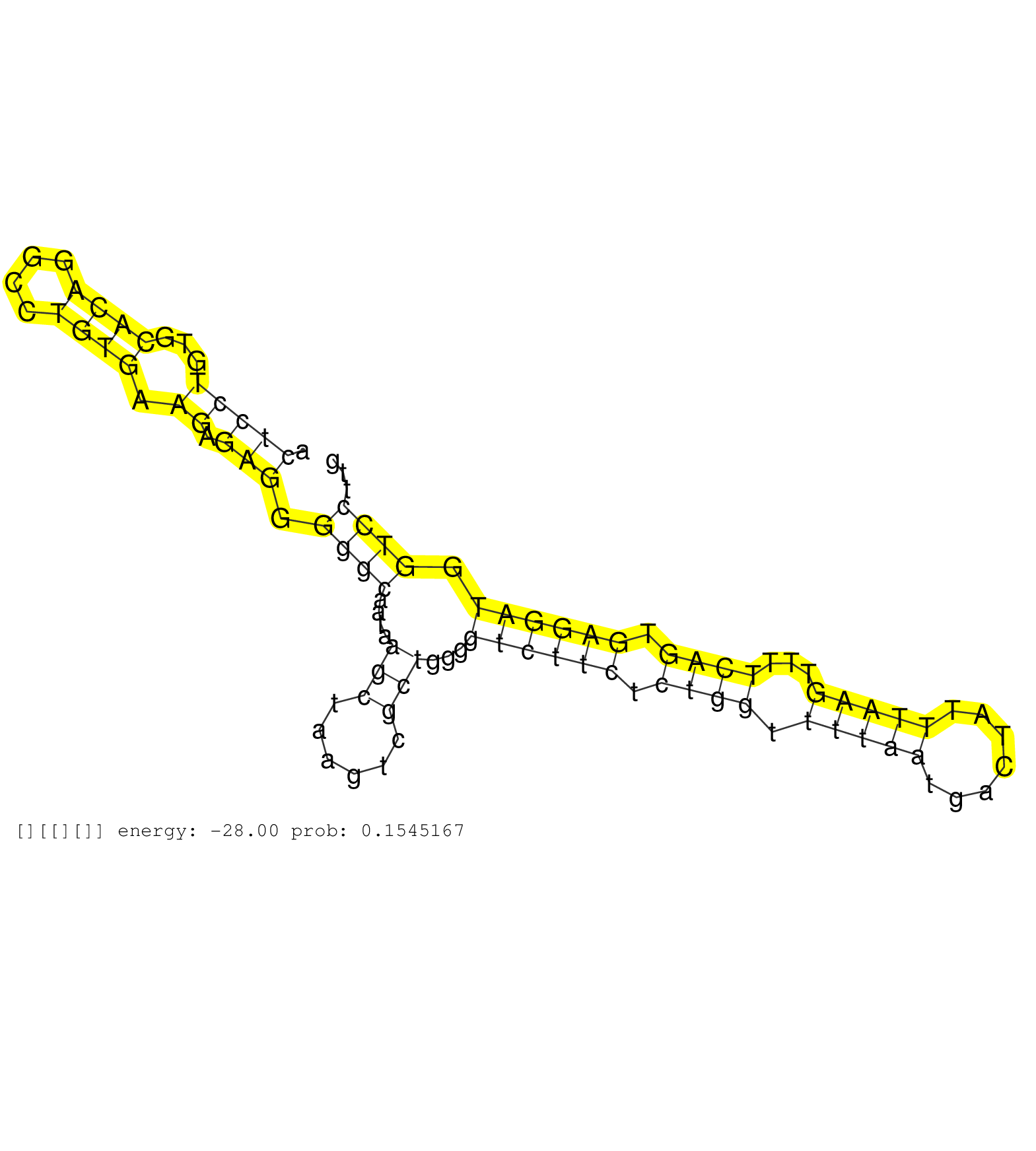

| Gene: Zfp41 | ID: uc007wgy.1_intron_0_0_chr15_75447459_f.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| CGCCTTATCCTGCCGGCTCCAGCAGCTCCTTGATGGACCTGAGTCCCTAGGTCAGCGGAATTCAACTAAGTTGCCTGAATCACTCGCACCTACTCCTGTGCACAGGCCTGTGAAGAGAGGGGGCAATAAGCTAAGTCGCTGGGGTCTTCTCTGGTTTTAATGACTATTTAAGTTTTCAGTGAGGATGGTCCTTGGTGAGCAGACACAGACACCTGGGTCCCGCTTGTCATTCCTCTCACCAGAGTGATGG ............................................................................................(((((...((((....)))).)).))).((((....(((......)))...((((((.((((.(((((.......)))))...)))).)))))).))))........................................................... ...........................................................................................92....................................................................................................194...................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................TGTGCACAGGCCTGTGAAGAGAGGG................................................................................................................................. | 25 | 1 | 5.00 | 5.00 | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................TGTGCACAGGCCTGTGAAGAGAGGGG................................................................................................................................ | 26 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TGTGCACAGGCCTGTGAAGAGAGG.................................................................................................................................. | 24 | 1 | 3.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................ACAGGCCTGTGAAGAGAGGGGGCAAT........................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................CTATTTAAGTTTTCAGTGAGGATGGTC............................................................ | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TGAAGAGAGGGGGCAATAAGCTAAGTC................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TAGGTCAGCGGAATTCAACTAAGTTGC................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................................TGTGCACAGGCCTGTGAAGAGAGGa................................................................................................................................. | 25 | a | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TCCTGCCGGCTCCAGCAGCTCCTTG.......................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................TAGGTCAGCGGAATTCAACTAAGTTGCCT.............................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........TGCCGGCTCCAGCAGCTCCTTGATGGA..................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TACTCCTGTGCACAGGCCTGTGAAGA...................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTCAGCGGAATTCAACTAAGTTGCCT.............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................TGTGCACAGGCCTGTGAAGAGAGGGa................................................................................................................................ | 26 | a | 1.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGGGTCCCGCTTGTCATTCCTCTC............. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................................GTTTTAATGACTATTTAAGTTTT.......................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................TGAATCACTCGCACCTACTCCTGTGCACAGt................................................................................................................................................ | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................TTCAACTAAGTTGCCTGAATCACTCGC................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGAGGATGGTCCTTGGTGAGCAGAC.............................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................TTGGTGAGCAGACACAGACACCTGGGTCCC............................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................TAGGTCAGCGGAATTCAACTAAGTTGa................................................................................................................................................................................ | 27 | a | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TGAGTCCCTAGGTCAGCGGAATTCAAC........................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGACTATTTAAGTTTTCAGTGAGGATG............................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................GTGAGGATGGTCCTTGGTGAGCc................................................. | 23 | c | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TGAAGAGAGGGGGCAATAAGCTAAa................................................................................................................... | 25 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGCCGGCTCCAGCAGCTCCTTGATGGACC................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGCCGGCTCCAGCAGCTCCTTGATGGACa................................................................................................................................................................................................................... | 29 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGACTATTTAAGTTTTCAGTGAGGA................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................................................TTTCAGTGAGGATGGTCCTTGGTG..................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TAGGTCAGCGGAATTCAACTAAGTTG................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................TTGATGGACCTGAGTCCCTAGGTCAGCGG................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| CGCCTTATCCTGCCGGCTCCAGCAGCTCCTTGATGGACCTGAGTCCCTAGGTCAGCGGAATTCAACTAAGTTGCCTGAATCACTCGCACCTACTCCTGTGCACAGGCCTGTGAAGAGAGGGGGCAATAAGCTAAGTCGCTGGGGTCTTCTCTGGTTTTAATGACTATTTAAGTTTTCAGTGAGGATGGTCCTTGGTGAGCAGACACAGACACCTGGGTCCCGCTTGTCATTCCTCTCACCAGAGTGATGG ............................................................................................(((((...((((....)))).)).))).((((....(((......)))...((((((.((((.(((((.......)))))...)))).)))))).))))........................................................... ...........................................................................................92....................................................................................................194...................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................TCTCTGGTTTTAATGAcc....................................................................................... | 18 | cc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |