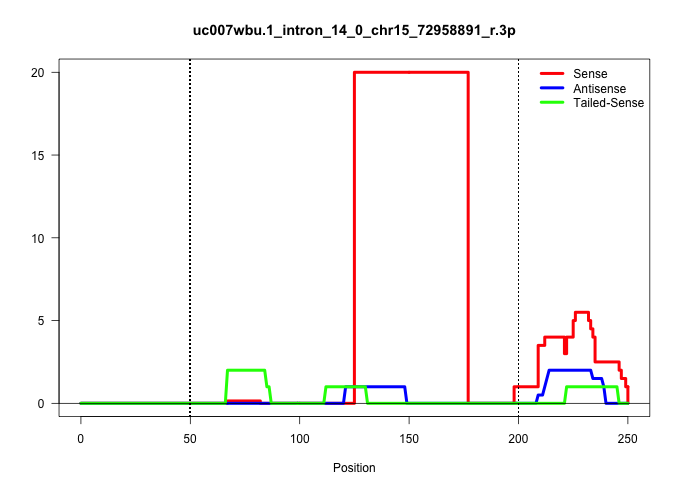
| Gene: Eif2c2 | ID: uc007wbu.1_intron_14_0_chr15_72958891_r.3p | SPECIES: mm9 |
 |
|
(5) OTHER.mut |
(5) PIWI.ip |
(17) TESTES |
| CTGCATTCTGTCAGTTCCTATGTTTAAACATCTGATAGTCTTAAGTCCACGGGGTTACTGCTTCCACAGGGAATGCAGGCTAGAACTACATGTAATGAGCATCTACAGAAGAAACCTAGAGCAGTCTCTGAGTGTAGCTATGCCTGCTATGATGATGGGCTGATTCCATCTGATTAAAAGAGGCCTTATCCCTGTTGCAGGTACACCCCTGTTGGCCGTTCCTTCTTCACTGCATCTGAAGGCTGTTCCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................CTCTGAGTGTAGCTATGCCTGCTATGATGATGGGCTGATTCCATCTGATTAA......................................................................... | 52 | 1 | 20.00 | 20.00 | 20.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................AGGGAATGCAGGCTAGta..................................................................................................................................................................... | 18 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TTCTTCACTGCATCTGAAGGatgt.... | 24 | atgt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGTTGGCCGTTCCTTCTTCACTGCAT............... | 26 | 2 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................AACCTAGAGCAGTCTaaaa....................................................................................................................... | 19 | aaaa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................AGGGAATGCAGGCTAGgatt................................................................................................................................................................... | 20 | gatt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGTACACCCCTGTTGGCCGTTC............................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TTCACTGCATCTGAAGGCTGTTCC. | 24 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGTTGGCCGTTCCTTCTTCACTGCA................ | 25 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TGGCCGTTCCTTCTTCACTGCAT............... | 23 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGTTGGCCGTTCCTTCTTCACTG.................. | 23 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ..................................................................................................................................................................................................................................TCACTGCATCTGAAGGCTGTTCCA | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ..............................................................................................................................................................................................................................TTCTTCACTGCATCTGAAGGCTGTT... | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TTCACTGCATCTGAAGGCTGTTCCA | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| .................................................................................................................................................................................................................TGTTGGCCGTTCCTTCTTCACTGC................. | 24 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TTCTTCACTGCATCTGAAGGCTGT.... | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................AGGGAATGCAGGCTA........................................................................................................................................................................ | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| CTGCATTCTGTCAGTTCCTATGTTTAAACATCTGATAGTCTTAAGTCCACGGGGTTACTGCTTCCACAGGGAATGCAGGCTAGAACTACATGTAATGAGCATCTACAGAAGAAACCTAGAGCAGTCTCTGAGTGTAGCTATGCCTGCTATGATGATGGGCTGATTCCATCTGATTAAAAGAGGCCTTATCCCTGTTGCAGGTACACCCCTGTTGGCCGTTCCTTCTTCACTGCATCTGAAGGCTGTTCCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................CTGCTATGATGATGccca............................................................................................. | 18 | ccca | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................CAGTCTCTGAGTGTAGCTATGCCTGCTA..................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................AGAGGCCTTATCCCTga......................................................... | 17 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TGGCCGTTCCTTCTTCACTGCATCTGA........... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................GCCGTTCCTTCTTCACTGCATCTGAA.......... | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| .....................................................................................................................................................................................................................GGCCGTTCCTTCTTCACTGCATCTGAA.......... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| .................................................................................................................................................................................................................TGTTGGCCGTTCCTTCTTCACTGCA................ | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |