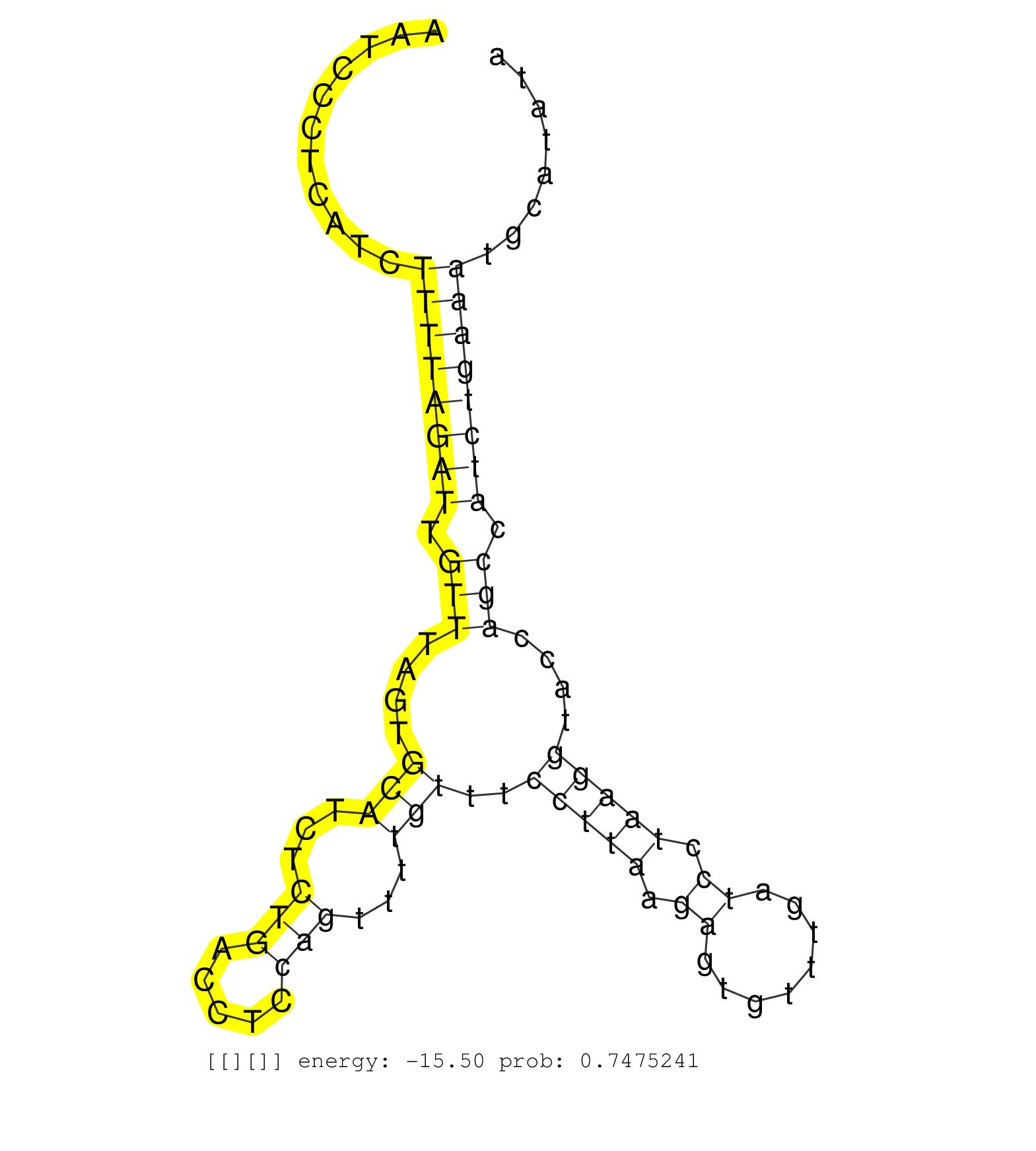
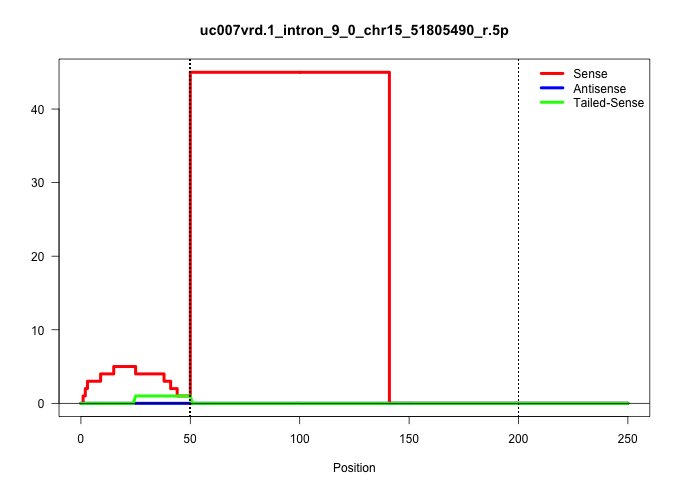
| Gene: Rad21 | ID: uc007vrd.1_intron_9_0_chr15_51805490_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
| ACTTTACCTGAGGAATTCCACGATTTTGATCAGCCACTGCCAGATTTAGAGTATGTCTGTCTTGTGTCTCTCCTCCCTTCTCAGTCCCTTCTTCTGCCCTAATCCCTCATCTTTTAGATTGTTTAGTGCATCTCTGACCTCCAGTTTTGTTTCCTTAAGAGTGTTTGATCCTAAGGTACCAGCCATCTGAAATGCATATACTTTGATTTAGGAGTATGTTCATTACAAGGTGTACGATTTAGGAAGTGTT ...............................................................................................................((((((((.(((....(((...(((.....)))...)))..(((((.((........)).)))))....))).)))))))).......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTATGTCTGTCTTGTGTCTCTCCTCCCTTCTCAGTCCCTTCTTCTGCCCTAA.................................................................................................................................................... | 52 | 1 | 49.00 | 49.00 | 49.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................TTTGATCAGCCACTGCCAGATTTAGA........................................................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| .CTTTACCTGAGGAATTCCACGATT................................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................TTTTGATCAGCCACTGCC................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...TTACCTGAGGAATTCCACGA................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..TTTACCTGAGGAATTCCACGAT.................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........GAGGAATTCCACGATTTTGATCAGCCACT.................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................TTGATCAGCCACTGCCAGATTTAGAt....................................................................................................................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............TTCCACGATTTTGATCAGCCACTGCCAGA.............................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ACTTTACCTGAGGAATTCCACGATTTTGATCAGCCACTGCCAGATTTAGAGTATGTCTGTCTTGTGTCTCTCCTCCCTTCTCAGTCCCTTCTTCTGCCCTAATCCCTCATCTTTTAGATTGTTTAGTGCATCTCTGACCTCCAGTTTTGTTTCCTTAAGAGTGTTTGATCCTAAGGTACCAGCCATCTGAAATGCATATACTTTGATTTAGGAGTATGTTCATTACAAGGTGTACGATTTAGGAAGTGTT ...............................................................................................................((((((((.(((....(((...(((.....)))...)))..(((((.((........)).)))))....))).)))))))).......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................TATGTCTGTCTTGTGaaaa........................................................................................................................................................................................ | 19 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................CCTCCCTTCTCAGTCCCT................................................................................................................................................................. | 18 | 8 | 0.62 | 0.62 | - | - | - | - | - | - | - | - | - | 0.38 | 0.25 | - |
| ...................................................................CCTCCCTTCTCAGTCCCTgatt................................................................................................................................................................. | 22 | gatt | 0.12 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 0.12 |
| .......................................................................CCTCCCTTCTCAGTCCC.................................................................................................................................................................. | 17 | 12 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | 0.08 | - |