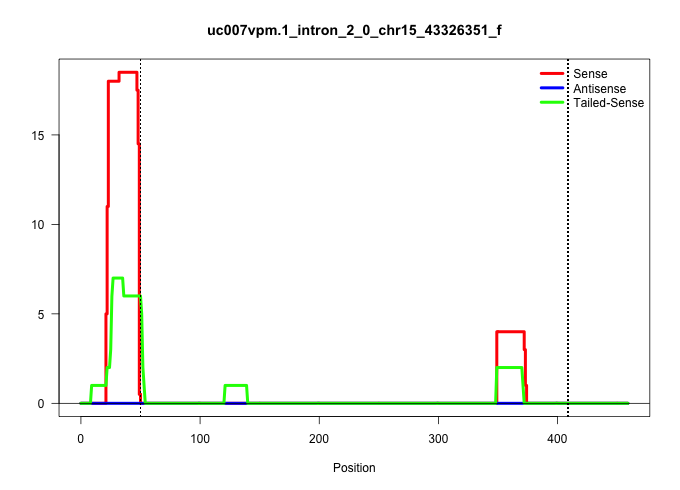
| Gene: Ttc35 | ID: uc007vpm.1_intron_2_0_chr15_43326351_f | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(3) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| AGCAGGTCATGATTGCAGCCCTAGACTATGGTCGGGATGACTTGGCATTGGTAAGTACTTAAATTAACTGTATTTTACTCTGTATTTGTTTTATGAGTTATAAATTAGGAAGTAGTTAAGGTTTGCTGTGGGGCATGTACTGAGTGCTTTCTGTAGCAAGTCTTGAATGCTTTTCTAAGTACGAGACTAGCCTGTAACTCAGTGGCAGGATGCTTGGCTAGCTTGTACAAGGTTCTGGATTTACTCTCTAGCATCATGAACACAAATTAAATAAATAAAATACCCATACCAATAAAATGTTAAGTGAAACTCTCATTAGAAACTCATTAGACCATTTTTAATGGATTATTTTGAAGATGTAATGGTGGTTGCTCAAGTCGTTATTTTGTTTAACCCATTATGTTTTCAGTTTTGTCTTCAGGAATTAAGAAGACAATTCCCTGGTAGTCACAGAGTTAA ......................................................................................................................................................................................................................................................................................................................((.(((((((((.((....))...)))))))))))......(((((((((((((..(((((..((((.......))))..))))))))))))))))))................................................... .....................................................................................................................................................................................................................................................................................................................310................................................................................................409................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................GACTATGGTCGGGATGACTTGGCATT.......................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 7.00 | 7.00 | 1.00 | 1.00 | 2.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ......................AGACTATGGTCGGGATGACTTGGCATT.......................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................TAGACTATGGTCGGGATGACTTGGCATT.......................................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................TATGGTCGGGATGACTTGGCATTGtt....................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | tt | 3.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................TTTGAAGATGTAATGGTGGTTGCT...................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................AGACTATGGTCGGGATGACTTGGCAT........................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................ATGGTCGGGATGACTTGGCATTGtttt..................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | tttt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................TTTGAAGATGTAATGGTGGgtgc....................................................................................... | 23 | gtgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................TTTGAAGATGTAATGGTGGTTGC....................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................TAGACTATGGTCGGGATGACTTGGCAT........................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................AGACTATGGTCGGGATGACTTGGCATTGt........................................................................................................................................................................................................................................................................................................................................................................................................................ | 29 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................CTATGGTCGGGATGACTTGGCATTGttt...................................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | ttt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................TTTGAAGATGTAATGGTGGTTGCTC..................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................TTTGAAGATGTAATGGTGttgc........................................................................................ | 22 | ttgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................TAGACTATGGTCGGGATGACTTGGCA............................................................................................................................................................................................................................................................................................................................................................................................................................ | 26 | 2 | 1.00 | 1.00 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TTTGCTGTGGGGCATagga............................................................................................................................................................................................................................................................................................................................... | 19 | agga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........TGATTGCAGCCCTAGACTATGGTCGGt....................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................CGGGATGACTTGGCATTG......................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| AGCAGGTCATGATTGCAGCCCTAGACTATGGTCGGGATGACTTGGCATTGGTAAGTACTTAAATTAACTGTATTTTACTCTGTATTTGTTTTATGAGTTATAAATTAGGAAGTAGTTAAGGTTTGCTGTGGGGCATGTACTGAGTGCTTTCTGTAGCAAGTCTTGAATGCTTTTCTAAGTACGAGACTAGCCTGTAACTCAGTGGCAGGATGCTTGGCTAGCTTGTACAAGGTTCTGGATTTACTCTCTAGCATCATGAACACAAATTAAATAAATAAAATACCCATACCAATAAAATGTTAAGTGAAACTCTCATTAGAAACTCATTAGACCATTTTTAATGGATTATTTTGAAGATGTAATGGTGGTTGCTCAAGTCGTTATTTTGTTTAACCCATTATGTTTTCAGTTTTGTCTTCAGGAATTAAGAAGACAATTCCCTGGTAGTCACAGAGTTAA ......................................................................................................................................................................................................................................................................................................................((.(((((((((.((....))...)))))))))))......(((((((((((((..(((((..((((.......))))..))))))))))))))))))................................................... .....................................................................................................................................................................................................................................................................................................................310................................................................................................409................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................TGACTTGGCATTGgcac......................................................................................................................................................................................................................................................................................................................................................................................................................... | 17 | gcac | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................GTATTTGTTTTATGtgc............................................................................................................................................................................................................................................................................................................................................................................ | 17 | tgc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................AACTCATTAGACCATgaac............................................................................................................................ | 19 | gaac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................................ACGAGACTAGCCTgatc.......................................................................................................................................................................................................................................................................... | 17 | gatc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |