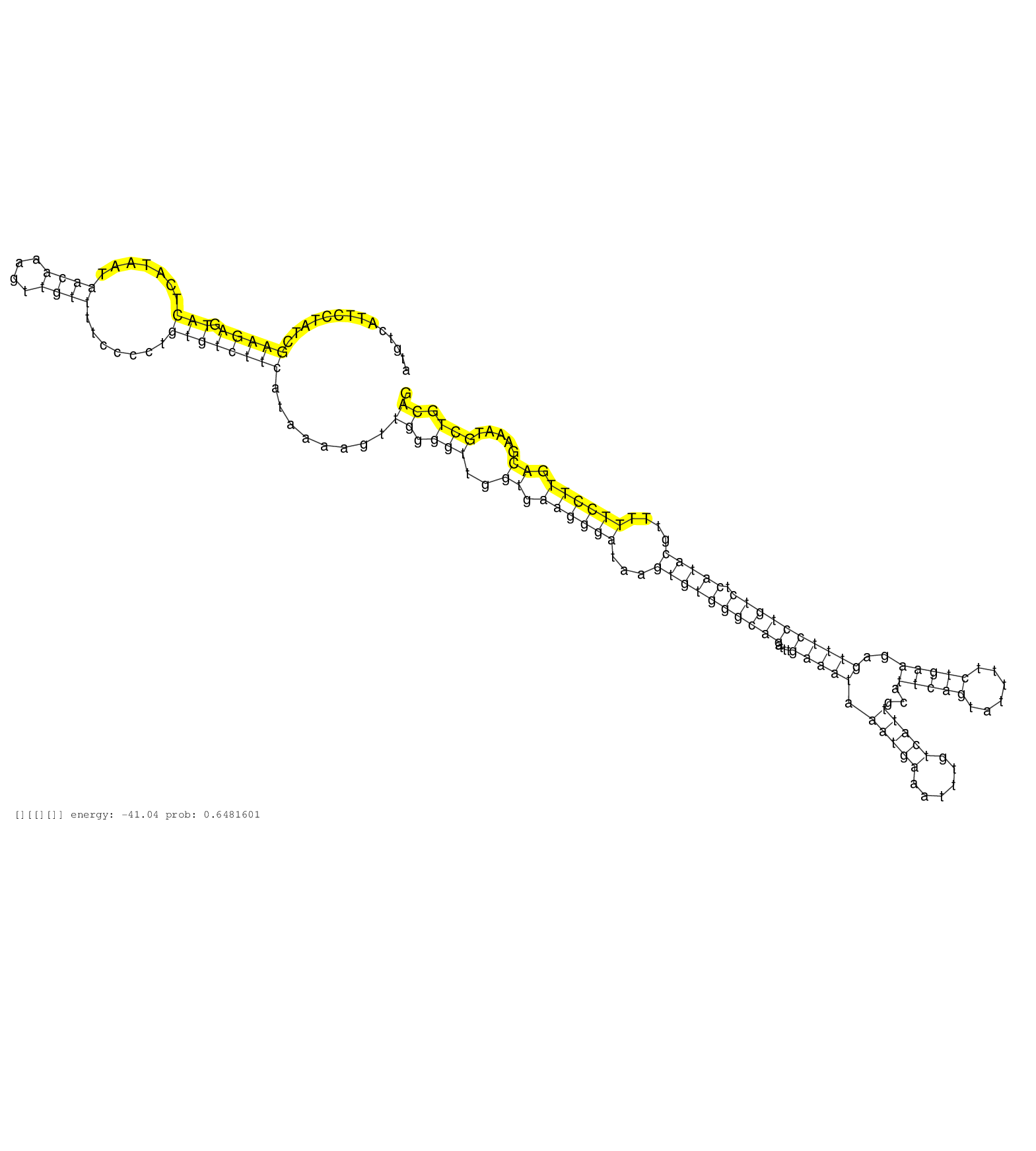

| Gene: March6 | ID: uc007vkh.1_intron_13_0_chr15_31415906_r | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(23) TESTES |
| CAACGATTGTTGGATATATACTTTTAGCAATAACACTGATAATTTGTCATGTATCCTTTAATGAACCAGCCATGTCATTCCTATCGAAGAGTACTCATAATAACAAAGTTGTTTTCCCCTGTGTCTTCATAAAAGTTGGGGTTGGTGAAGGGATAAGTGTGGGCAGATTTGAAATAAATGAAATTTGTCATTGCATTCAGTATTTTCTGAAGAGTTTCCTGTCTCATACGTTTTTCCTTGACGAAATGCTGCAGGCATTGGCAACTCTTGTTAAATTTCATAGGTCTCGTCGCTTGCTGGGCGT .....................................................................................(((((.(((.......((((....)))).......))))))))........((.(((..((.((((((...((((((((((....(((((.(((((......)))))...(((((......)))))..)))))))))).)))))....)))))).)).....))).))................................................... .......................................................................72....................................................................................................................................................................................254................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................TTTTCCTTGACGAAATGCTGCAG.................................................. | 23 | 1 | 10.00 | 10.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - |
| ..................................................................................................................................................................................................................................TACGTTTTTCCTTGACGAAATGCTGCAG.................................................. | 28 | 1 | 5.00 | 5.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................TTCCTTGACGAAATGCTGCAG.................................................. | 21 | 1 | 4.00 | 4.00 | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................ACGTTTTTCCTTGACGAAATGCTGCAG.................................................. | 27 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................TAAATTTCATAGGTCTCGTCGCTTGCT...... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................ATACGTTTTTCCTTGACGAAATGCTGCAG.................................................. | 29 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................TTCCTTGACGAAATGCTGCAGa................................................. | 22 | a | 3.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - |
| ...........................................................................................................................................................................................................................................CCTTGACGAAATGCTGCAG.................................................. | 19 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................TTTCCTTGACGAAATGCTGCAGa................................................. | 23 | a | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................CTCTTGTTAAATTTCATAGGTCTCGTCGC........... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .AACGATTGTTGGATATATACTTTTAGCA................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TGGATATATACTTTTAGCAATAACACTGAT........................................................................................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................TCCTTGACGAAATGCTGCAG.................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................TTTTTCCTTGACGAAATGCTGCAG.................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................TTCCTTGACGAAATGCTGCAGaa................................................ | 23 | aa | 1.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................TAAATTTCATAGGTCTCGTCGCTTGCTG..... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................AAATTTCATAGGTCTCGTCGCTTGCTGGG... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................ATTCCTATCGAAGAGTACTCATAAT........................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................TTTCCTTGACGAAATGCTGCA................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................AGGTCTCGTCGCTTGCTG..... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................CTTGACGAAATGCTGCAG.................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................................................................................................................................................................................TTTCCTTGACGAAATGCTGCAGaa................................................ | 24 | aa | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............ATATATACTTTTAGCAATAACACTGATAATTT................................................................................................................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................AATTTGTCATGTATCCTTTAATGAACCAGC.......................................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........TGGATATATACTTTTAGC.................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| CAACGATTGTTGGATATATACTTTTAGCAATAACACTGATAATTTGTCATGTATCCTTTAATGAACCAGCCATGTCATTCCTATCGAAGAGTACTCATAATAACAAAGTTGTTTTCCCCTGTGTCTTCATAAAAGTTGGGGTTGGTGAAGGGATAAGTGTGGGCAGATTTGAAATAAATGAAATTTGTCATTGCATTCAGTATTTTCTGAAGAGTTTCCTGTCTCATACGTTTTTCCTTGACGAAATGCTGCAGGCATTGGCAACTCTTGTTAAATTTCATAGGTCTCGTCGCTTGCTGGGCGT .....................................................................................(((((.(((.......((((....)))).......))))))))........((.(((..((.((((((...((((((((((....(((((.(((((......)))))...(((((......)))))..)))))))))).)))))....)))))).)).....))).))................................................... .......................................................................72....................................................................................................................................................................................254................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|