
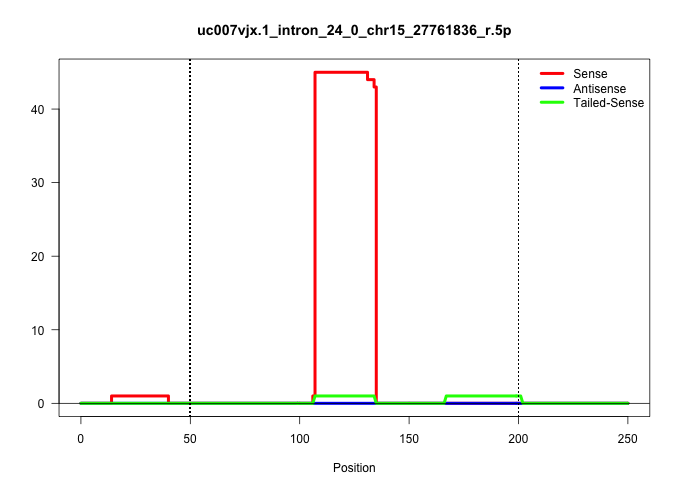
| Gene: Trio | ID: uc007vjx.1_intron_24_0_chr15_27761836_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(14) TESTES |
| GCTGCCCACGAGCTTAATGAAGAGAAGCGGAAGTCTGCCCGCAGGAAAGAGTAAGCCAGCATTCCCTGCAATGGTGAGGGGAGCGTCAGAGCCCACAGACAGGCTGCTGCCATGATGTCAGTCAAGTGTCTGAGACCTCAGTTGGCTTTGTCGGTTGACTTCTCTCTGTGCTGACCCCTGATCACATGGCTCACTGCCTTGCCCTGGTTTCTCTATGCTTTCCTTTCCTGGTGTGACAAATTGCTTGCCA .............................................................................................................(((((..(((((((.(((....((((..((((..(((...)))..))))..)))).....))))))...))))..)))))............................................................. ......................................................................................................103...................................................................................189........................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................TGCCATGATGTCAGTCAAGTGTCTGAGA................................................................................................................... | 28 | 1 | 45.00 | 45.00 | 12.00 | 9.00 | 3.00 | 6.00 | 5.00 | 4.00 | 5.00 | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................CAAGTGTCTGAGACCTCAGTTGGC........................................................................................................ | 24 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................CAAGTGTCTGAGACCTCAGTTGtc........................................................................................................ | 24 | tc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGCCATGATGTCAGTCAAGTGTCTGAG.................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................GCCATGATGTCAGTCAAGTGTCTGAGA................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................TGCCATGATGTCAGTCAAGTGTCTGgga................................................................................................................... | 28 | gga | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................GTGCTGACCCCTGAaaag................................................................. | 18 | aaag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................CTGCCATGATGTCAGTCAAGTGTCTGAGA................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................................................................................ATGGCTCACTGCCgttt................................................ | 17 | gttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............TAATGAAGAGAAGCGGAAGTCTGCCC.................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................TGCCATGATGTCAGTCAAGTGTCT....................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| GCTGCCCACGAGCTTAATGAAGAGAAGCGGAAGTCTGCCCGCAGGAAAGAGTAAGCCAGCATTCCCTGCAATGGTGAGGGGAGCGTCAGAGCCCACAGACAGGCTGCTGCCATGATGTCAGTCAAGTGTCTGAGACCTCAGTTGGCTTTGTCGGTTGACTTCTCTCTGTGCTGACCCCTGATCACATGGCTCACTGCCTTGCCCTGGTTTCTCTATGCTTTCCTTTCCTGGTGTGACAAATTGCTTGCCA .............................................................................................................(((((..(((((((.(((....((((..((((..(((...)))..))))..)))).....))))))...))))..)))))............................................................. ......................................................................................................103...................................................................................189........................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|