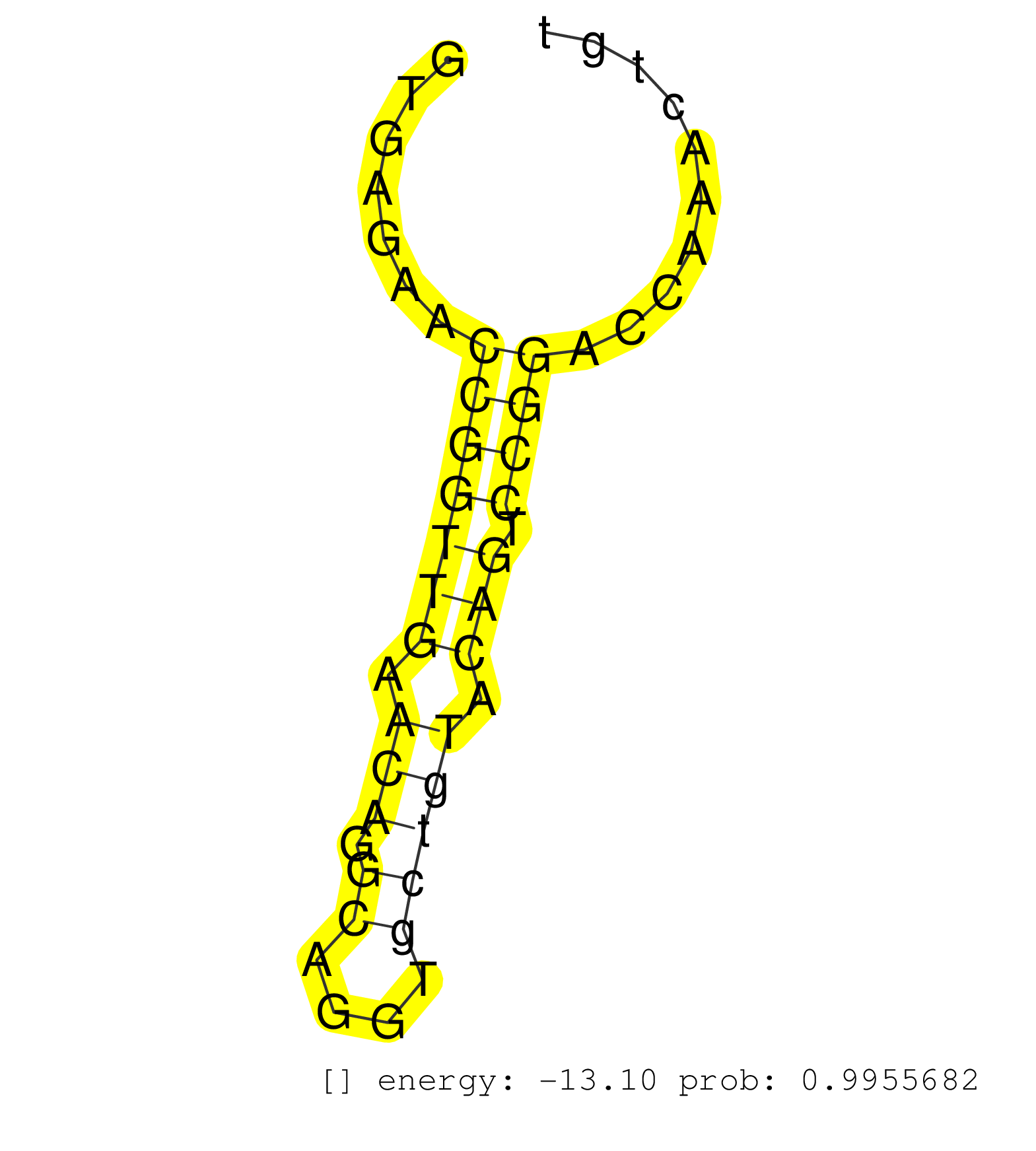

| Gene: Myo10 | ID: uc007vjc.1_intron_38_0_chr15_25737944_f.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) PIWI.ip |
(11) TESTES |
| CCCTGATCAAGGAGTGGCCTGGTTATGGATCAACACTCTTTGATGTGGAGGTGAGAACCGGTTGAACAGGCAGGTGCTGTACAGTCCGGACCAAACTGTCTCAGAACCTGAACCTCCCTGGGGTCACACAAAGGGACAGAGGGACCACTTGGCTCTGTGCCTTCTGAGTCTCACCACGAGCATCCTCGTGTCCCACACGGGTTTTGTTTTGTTTTTTTGTTGTGTTTTCTTCGGTAGTTTTTGTTTTTAC .........................................................(((((((.(((.((....))))).))).))))................................................................................................................................................................. ..................................................51..............................................99...................................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT2() Testes Data. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGAACCGGTTGAACAGGCAGGT............................................................................................................................................................................... | 25 | 1 | 19.00 | 19.00 | 15.00 | 4.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................TACAGTCCGGACgatc........................................................................................................................................................... | 16 | gatc | 3.00 | 0.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTGAGAACCGGTTGAACAGGCAGGTG.............................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAACCGGTTGAACAGGCAGG................................................................................................................................................................................ | 24 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTGAGAACCGGTTGAACAGGCAGGg............................................................................................................................................................................... | 25 | g | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................GCTGTACAGTCCGGACCAAACTGTCTCAGA................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................................................................CCACACGGGTTTTGTTTTGTTTTTgag............................... | 27 | gag | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAACCGGTTGAACAGGCAGGTt.............................................................................................................................................................................. | 26 | t | 1.00 | 19.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGTGCCTTCTGAGTCTCACCACGAGC..................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGTTTTCTTCGGTAGTTTTTGTTTT... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTGAGAACCGGTTGAACAG..................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGAACCGGTTGAACAGGCAGGTG.............................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................GAGAACCGGTTGAACAGGCAGGTGC............................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...TGATCAAGGAGTGGCCcct.................................................................................................................................................................................................................................... | 19 | cct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................TATGGATCAACACTCTTTGATGTGGA......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAACCGGTTGAACAGGCAGGTa.............................................................................................................................................................................. | 26 | a | 1.00 | 19.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAACCGGTTGAACAGGCAGGTGC............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......TCAAGGAGTGGCCTGGTTATt............................................................................................................................................................................................................................... | 21 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| CCCTGATCAAGGAGTGGCCTGGTTATGGATCAACACTCTTTGATGTGGAGGTGAGAACCGGTTGAACAGGCAGGTGCTGTACAGTCCGGACCAAACTGTCTCAGAACCTGAACCTCCCTGGGGTCACACAAAGGGACAGAGGGACCACTTGGCTCTGTGCCTTCTGAGTCTCACCACGAGCATCCTCGTGTCCCACACGGGTTTTGTTTTGTTTTTTTGTTGTGTTTTCTTCGGTAGTTTTTGTTTTTAC .........................................................(((((((.(((.((....))))).))).))))................................................................................................................................................................. ..................................................51..............................................99...................................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT2() Testes Data. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................TGGCTCTGTGCCTTCTGAGTCTCACC........................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................GGACCAAACTGTCTCcta.................................................................................................................................................... | 18 | cta | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |