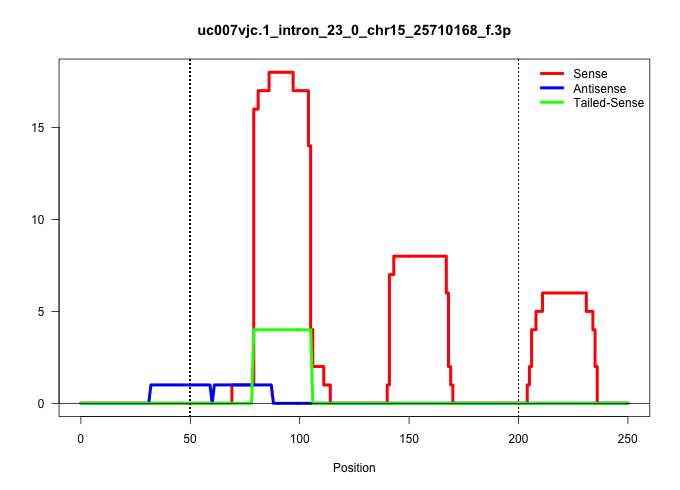
| Gene: Myo10 | ID: uc007vjc.1_intron_23_0_chr15_25710168_f.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
| AGATTGGATGCAGGCTCCAGTCATCGGGCCCCACCTCATTTTAACTTAGTCACCCGATTAAAAGCCCCTTGTCCAACCGTAGTAGTTATTCTGAGGCGCTGGGGTTAGGGTTTCAGCTTGTCGTTTGGGTGGGGCCACAGTTCAGTGTGTAGAACAGGAAGAAGGTGCTTGTTCTAAAACCCACCTTGCCCCTGGTCCAGGAGGCAGAGACAAGGAGGCAGCAAGAACTGGAAGCCTTGCAGAAGAGCCA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................TAGTAGTTATTCTGAGGCGCTGGGGT................................................................................................................................................. | 26 | 1 | 10.00 | 10.00 | 5.00 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................TAGTAGTTATTCTGAGGCGCTGGGGTa................................................................................................................................................ | 27 | a | 4.00 | 10.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TCAGTGTGTAGAACAGGAAGAAGGTGC.................................................................................. | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 |
| ...............................................................................TAGTAGTTATTCTGAGGCGCTGGGG.................................................................................................................................................. | 25 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................TCAGTGTGTAGAACAGGAAGAAGGTG................................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................TAGTAGTTATTCTGAGGCGCTGGGGTT................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................GTAGTTATTCTGAGGCGCTGGGGTTAGGGTTTC........................................................................................................................................ | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................AGTGTGTAGAACAGGAAGAAGGTGCT................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................GACAAGGAGGCAGCAAGAACTGGAAGC............... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................................................................GAGACAAGGAGGCAGCAAGAACTGG................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AAGGAGGCAGCAAGAACTGGAAGC............... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................GAGACAAGGAGGCAGCAAGAACTGGAAG................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TGTCCAACCGTAGTAGTTATTCTGAGGC......................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGAGACAAGGAGGCAGCAAGAACTGGAAGCC.............. | 31 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TCAGTGTGTAGAACAGGAAGAAGGTGCTT................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TTCAGTGTGTAGAACAGGAAGAAGGTGC.................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TATTCTGAGGCGCTGGGGTTAGGGT........................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................CAGAGACAAGGAGGCAGCAAGAACTGGAAGCC.............. | 32 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGATTGGATGCAGGCTCCAGTCATCGGGCCCCACCTCATTTTAACTTAGTCACCCGATTAAAAGCCCCTTGTCCAACCGTAGTAGTTATTCTGAGGCGCTGGGGTTAGGGTTTCAGCTTGTCGTTTGGGTGGGGCCACAGTTCAGTGTGTAGAACAGGAAGAAGGTGCTTGTTCTAAAACCCACCTTGCCCCTGGTCCAGGAGGCAGAGACAAGGAGGCAGCAAGAACTGGAAGCCTTGCAGAAGAGCCA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................CTTGTTCTAAAACCatgc..................................................................... | 18 | atgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................ACCTCATTTTAACTTAGTCACCCGATTA.............................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................AAGCCCCTTGTCCAACCGTAGTAGTTA.................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |