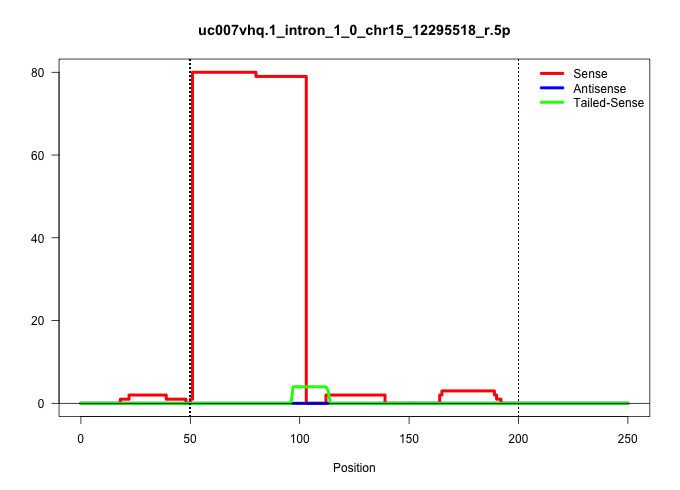
| Gene: Pdzd2 | ID: uc007vhq.1_intron_1_0_chr15_12295518_r.5p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(1) OVARY |
(1) PIWI.ip |
(10) TESTES |
| TGGGAAGGCCCCATTCCCCAGAAGGACCTTGCCGCTGGAGCCAGGAGCTGGTGAGTTTTATCCACTGATAGGGGCGAGGGGCGTGAGCCACTGGTTGATAAGGACTGTACCCTGGATTACTTTGTACTAGCAAGATAAAAAAAGGCTAGTGAATGAGATAGCTTTTGTAATGGGACTCGATGTGAAAGGCTTCTAAACAAGGCGTGTCCTTGAGAAACATCAAGATTTCTGTTTCCTGTTTTAACAGACT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................TGAGTTTTATCCACTGATAGGGGCGAGGGGCGTGAGCCACTGGTTGATAAGG................................................................................................................................................... | 52 | 1 | 79.00 | 79.00 | 79.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TGGATTACTTTGTACTAGCAAGATAAA............................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................ATAAGGACTGTACgact........................................................................................................................................ | 17 | gact | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................ATAAGGACTGTACgaca........................................................................................................................................ | 17 | gaca | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TTGTAATGGGACTCGATGTGAAAGGC............................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................ATAAGGACTGTACCac......................................................................................................................................... | 16 | ac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................AGGACCTTGCCGCTGGAGCCAGGAGC.......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................TTGTAATGGGACTCGATGTGAAAGG............................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTGAGTTTTATCCACTGATAGGGGCGAGGG.......................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TGTAATGGGACTCGATGTGAAAGGCTT.......................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................ATAAGGACTGTACCacc........................................................................................................................................ | 17 | acc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................CAGAAGGACCTTGCCGCTGGA................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| TGGGAAGGCCCCATTCCCCAGAAGGACCTTGCCGCTGGAGCCAGGAGCTGGTGAGTTTTATCCACTGATAGGGGCGAGGGGCGTGAGCCACTGGTTGATAAGGACTGTACCCTGGATTACTTTGTACTAGCAAGATAAAAAAAGGCTAGTGAATGAGATAGCTTTTGTAATGGGACTCGATGTGAAAGGCTTCTAAACAAGGCGTGTCCTTGAGAAACATCAAGATTTCTGTTTCCTGTTTTAACAGACT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................TACCCTGGATTACTggct................................................................................................................................. | 18 | ggct | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................TACCCTGGATTACTTggct................................................................................................................................ | 19 | ggct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................TACCCTGGATTACTTggca................................................................................................................................ | 19 | ggca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |