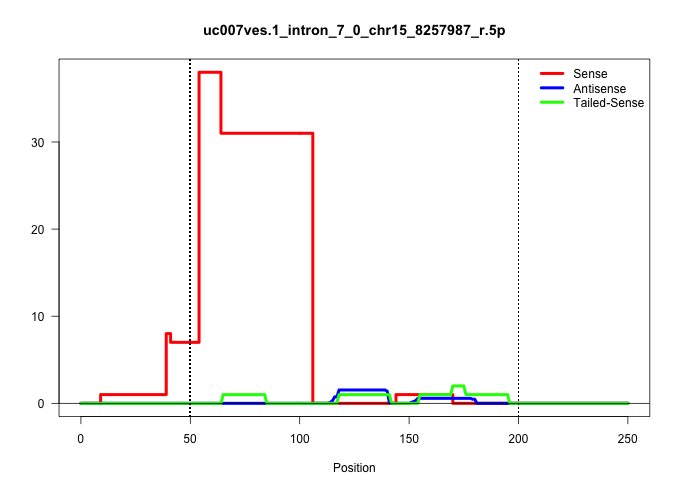
| Gene: Nipbl | ID: uc007ves.1_intron_7_0_chr15_8257987_r.5p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(7) PIWI.ip |
(23) TESTES |
| TGGTTAAACATGCCATGACTATGCAGCCGTACCTCACCACTAAGTGTAGTGTGAGTATAAGTGCTTTCTGTTGGTTGGCTGGTTTTCTGAGACAGGGTTTCTCTGTGCAGCGGCCCTGACTGTCCTGGACCTCACTCTGTAGACATTGGGCTGCTTCTGCCTCCCAAGTGCCGGGATTAAAGGCCTGCATGCACCACCACACCCCGCTTGTGCTGTCTTGTTTTTTTCTTTTCTTGCATGCAACAGGCAG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................GTATAAGTGCTTTCTGTTGGTTGGCTGGTTTTCTGAGACAGGGTTTCTCTGT................................................................................................................................................ | 52 | 1 | 31.00 | 31.00 | 31.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................CTAAGTGTAGTGTGAGTATAAGTGC.......................................................................................................................................................................................... | 25 | 1 | 7.00 | 7.00 | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................ATTGGGCTGCTTCTGCCTCCCAAGTG................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........ATGCCATGACTATGCAGCCGTACCTCACCACT................................................................................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TCTGCCTCCCAAGTGCCGagt.......................................................................... | 21 | agt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................ACTGTCCTGGACCTCACTCTattg............................................................................................................ | 24 | attg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TTCTGTTGGTTGGCTGaggc..................................................................................................................................................................... | 20 | aggc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................................................................................CCGGGATTAAAGGCCTGCATGCACCt...................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| TGGTTAAACATGCCATGACTATGCAGCCGTACCTCACCACTAAGTGTAGTGTGAGTATAAGTGCTTTCTGTTGGTTGGCTGGTTTTCTGAGACAGGGTTTCTCTGTGCAGCGGCCCTGACTGTCCTGGACCTCACTCTGTAGACATTGGGCTGCTTCTGCCTCCCAAGTGCCGGGATTAAAGGCCTGCATGCACCACCACACCCCGCTTGTGCTGTCTTGTTTTTTTCTTTTCTTGCATGCAACAGGCAG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................TGCCTCCCAAGTGCCGGGATTAAAtt..................................................................... | 26 | tt | 3.00 | 0.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TCCTGGACCTCACTCTGTAaagt............................................................................................................. | 23 | aagt | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................CCAAGTGCCGGGATTAAAGGat................................................................... | 22 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................GTCCTGGACCTCACTCTGTcg.............................................................................................................. | 21 | cg | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................CCTGGACCTCACTCTGTAGtttt............................................................................................................ | 23 | tttt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CTGTCCTGGACCTCACTCTGTAaga............................................................................................................. | 25 | aga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................CCTGGACCTCACTCTGTAtag............................................................................................................. | 21 | tag | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................GTCCTGGACCTCACTCTGTAcgc............................................................................................................. | 23 | cgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................CTGTCCTGGACCTCACTCTGTAGaaag............................................................................................................ | 27 | aaag | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................CTGCCTCCCAAGTGCCGGGATTAgtat....................................................................... | 27 | gtat | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GCCCTGACTGTCCTGGatc.......................................................................................................................... | 19 | atc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................CTGTCCTGGACCTCACTCTGTAacgg............................................................................................................. | 26 | acgg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................ACTGTCCTGGACCTCACTCTGTAGAta........................................................................................................... | 27 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGTCCTGGACCTCACTCTGTAcaca............................................................................................................. | 25 | caca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................CATGCACCACCACACCCgcc.............................................. | 20 | gcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGCCTCCCAAGTGCCGGGATTAtt....................................................................... | 24 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................CTGTCCTGGACCTCACTCTGTAaaa............................................................................................................. | 25 | aaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................ACTGTCCTGGACCTCACTCTGTAGAaaa........................................................................................................... | 28 | aaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................ACTGTCCTGGACCTCACTCTGTA............................................................................................................. | 23 | 14 | 0.71 | 0.71 | - | - | 0.21 | - | 0.21 | 0.07 | - | - | 0.07 | - | 0.07 | - | - | - | - | - | - | - | - | 0.07 | - | - | - |
| ....................................................................................................................TGACTGTCCTGGACCTCACTCTGTA............................................................................................................. | 25 | 14 | 0.29 | 0.29 | - | - | 0.14 | - | - | - | - | - | 0.07 | - | - | 0.07 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TTCTGCCTCCCAAGTGCCGGGATTAAA..................................................................... | 27 | 11 | 0.27 | 0.27 | - | - | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.09 | 0.09 |
| ...................................................................................................................CTGACTGTCCTGGACCTCACTCTGTA............................................................................................................. | 26 | 10 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................CCTGACTGTCCTGGACCTCACTCTGTA............................................................................................................. | 27 | 9 | 0.11 | 0.11 | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TGCTTCTGCCTCCCAAGTGCCGGGATTA....................................................................... | 28 | 10 | 0.10 | 0.10 | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................GCTTCTGCCTCCCAAGTGCCGGGATTAAA..................................................................... | 29 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................CTTCTGCCTCCCAAGTGCCGGGATTAAA..................................................................... | 28 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - |
| ....................................................................................................................TGACTGTCCTGGACCTCACTCTGTAG............................................................................................................ | 26 | 13 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................ACTGTCCTGGACCTCACTCTGTAaa............................................................................................................. | 25 | aa | 0.07 | 0.00 | - | - | - | - | - | - | - | - | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGACTGTCCTGGACCTCACTCTGT.............................................................................................................. | 24 | 14 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 | - | - | - |
| ......................................................................................................................ACTGTCCTGGACCTCACTCTGT.............................................................................................................. | 22 | 14 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 | - | - | - |
| ..................................................................................................................ACTGTCCTGGACCTCACTCTGTAaaat............................................................................................................. | 27 | aaat | 0.07 | 0.00 | - | - | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |