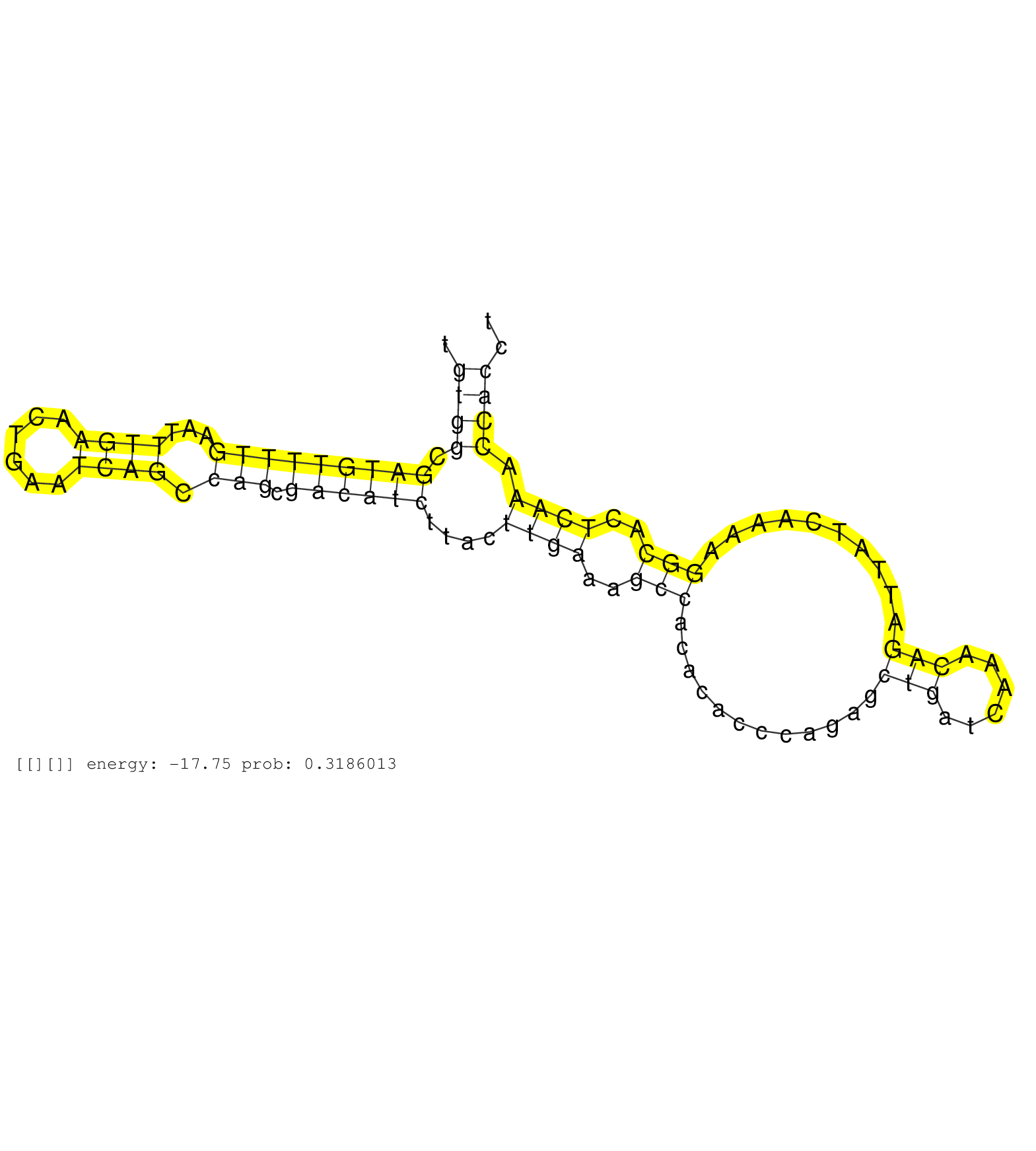
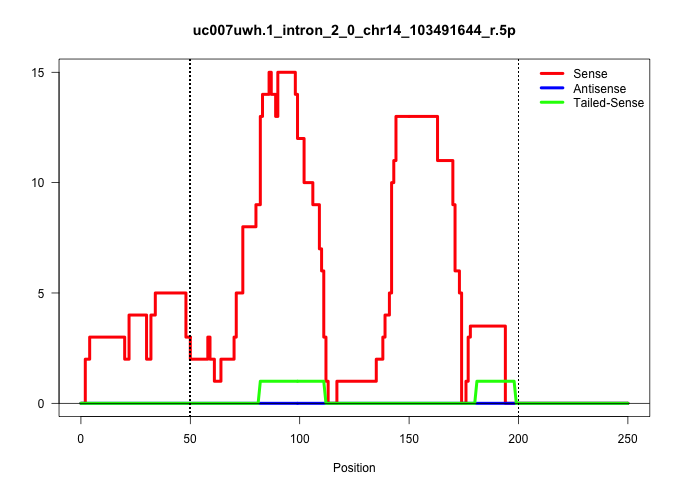
| Gene: Fbxl3 | ID: uc007uwh.1_intron_2_0_chr14_103491644_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(21) TESTES |
| CTCTTCTTGACCGGGCTCATGCTTCCCAAGTGTGCCGGAATTGGAATCAGGTATTTCACATGCCTGACTTGTGGCGATGTTTTGAATTTGAACTGAATCAGCCAGCGACATCTTACTTGAAAGCCACACACCCAGAGCTGATCAAACAGATTATCAAAAGGCACTCAAACCACCTACAGTATGTCAGCTTCAAGGTAACCCCTTCCATCCTTCTGTTAACGAGTAAAGCGCCTCACTGCCGTTACTGTCA ......................................................................((((.(((((((((...((((......)))).))).))))))....((((..(((............(((......)))..........)))..)))).))))............................................................................. .....................................................................70.......................................................................................................175......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................CAAACAGATTATCAAAAGGCACTCAAACC............................................................................... | 29 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................TGGCGATGTTTTGAATTTGAACTGAATC....................................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................CTGAATCAGCCAGCGACATCTTA....................................................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................CGATGTTTTGAATTTGAACTGAATCAGC.................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TTCCCAAGTGTGCCGGAATTGGAATC.......................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................AACAGATTATCAAAAGGCACTCAAACCACC............................................................................ | 30 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................AGTATGTCAGCTTCAAG........................................................ | 17 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TGTGGCGATGTTTTGAATTTGA............................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................AACTGAATCAGCCAGCGACATCT......................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..CTTCTTGACCGGGCTCATGCTTCCCAAG............................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGAATTTGAACTGAATCAGCCAGCGAC............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GACTTGTGGCGATGTTTTGAATTTGAAC............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGAATTTGAACTGAATCAGCCAGCGACATC.......................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................GAATTTGAACTGAATCAGCCAGCGACATC.......................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AGCTGATCAAACAGATTATCAAAAGGCA....................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TCAAACAGATTATCAAAAGGCACTCAAAC................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGAATTTGAACTGAATCAGCCAGCGACA............................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................GTGGCGATGTTTTGAATTTGAACTGAAT........................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TCTTGACCGGGCTCATGCTTCCCAAG............................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................AACTGAATCAGCCAGCGACAT........................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................GATCAAACAGATTATCAAAAGGCA....................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TGACTTGTGGCGATGTTTTGAATTT................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGAATTTGAACTGAATCAGCCAGCGACATt.......................................................................................................................................... | 30 | t | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CAGTATGTCAGCTTCAAG........................................................ | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TGTCAGCTTCAAGGTgga................................................... | 18 | gga | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................CCGGAATTGGAATCAG........................................................................................................................................................................................................ | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CAAACAGATTATCAAAAGGCACTCAAACCACC............................................................................ | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................AATTTGAACTGAATCAGCCAGCGACA............................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................TGAATTTGAACTGAATCAGCCAGC................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................AAACAGATTATCAAAAGGCACTCAAACCACC............................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................CATGCCTGACTTGTGGCGATGTTTTGAAT................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................ACAGATTATCAAAAGGCACTCAAACCACC............................................................................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..CTTCTTGACCGGGCTCAT...................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TTTGAATTTGAACTGAATCAGCCAGCGAC............................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TGCCGGAATTGGAATCAGGTATTTCACAT............................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................TGAAAGCCACACACCCAGAGCTGATCAA......................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CAAACAGATTATCAAAAGGCACTCAAACCAC............................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TGCCGGAATTGGAATCAGGTATTTCAC............................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TGATCAAACAGATTATCAAAAGGCACTCAAAC................................................................................ | 32 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................CGATGTTTTGAATTTGAACTGAATCAGCCAGC................................................................................................................................................ | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................TGAATTTGAACTGAATCAGCCAGCGACAT........................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TTTGAACTGAATCAGCCAGCGACAT........................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................GTATGTCAGCTTCAAG........................................................ | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| CTCTTCTTGACCGGGCTCATGCTTCCCAAGTGTGCCGGAATTGGAATCAGGTATTTCACATGCCTGACTTGTGGCGATGTTTTGAATTTGAACTGAATCAGCCAGCGACATCTTACTTGAAAGCCACACACCCAGAGCTGATCAAACAGATTATCAAAAGGCACTCAAACCACCTACAGTATGTCAGCTTCAAGGTAACCCCTTCCATCCTTCTGTTAACGAGTAAAGCGCCTCACTGCCGTTACTGTCA ......................................................................((((.(((((((((...((((......)))).))).))))))....((((..(((............(((......)))..........)))..)))).))))............................................................................. .....................................................................70.......................................................................................................175......................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|